3P9K
 
 | |
5YXT
 
 | |
6CL4
 
 | | LipC12 - Lipase from metagenomics | | Descriptor: | Lipase C12 | | Authors: | Iulek, J, Martini, V.P, Krieger, N, Glogauer, A, Souza, E.M. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure solution and analyses of the first true lipase obtained from metagenomics indicate potential for increased thermostability.
N Biotechnol, 53, 2019
|
|
6E6M
 
 | | Crystal structure of human cellular retinol-binding protein 1 in complex with cannabidiorcin (CBDO) | | Descriptor: | (1'R,2'R)-4,5'-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,6-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
5FYQ
 
 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
2X7X
 
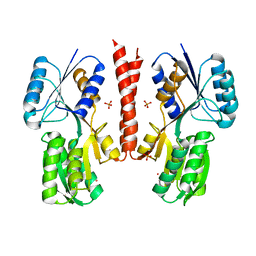 | | Fructose binding periplasmic domain of hybrid two component system BT1754 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, SENSOR PROTEIN, ... | | Authors: | Sonnenburg, E.D, Zheng, H, Joglekar, P, Higginbottom, S, Firbank, S.J, Bolam, D.N, Sonnenburg, J.L. | | Deposit date: | 2010-03-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations.
Cell(Cambridge,Mass.), 141, 2010
|
|
5FKA
 
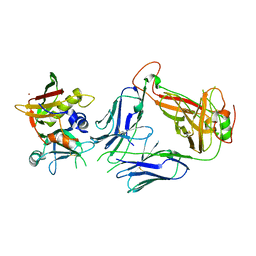 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN E, T CELL RECEPTOR ALPHA CHAIN, T CELL RECEPTOR BETA CHAIN, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins.
Sci.Rep., 6, 2016
|
|
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5Z51
 
 | |
2GIW
 
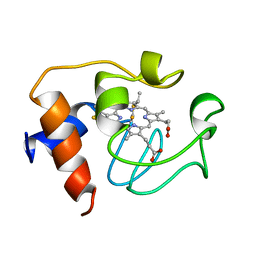 | | SOLUTION STRUCTURE OF REDUCED HORSE HEART CYTOCHROME C, NMR, 40 STRUCTURES | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Huber, J.G, Spyroulias, G.A, Turano, P. | | Deposit date: | 1998-06-25 | | Release date: | 1998-12-09 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced horse heart cytochrome c.
J.Biol.Inorg.Chem., 4, 1999
|
|
6EBE
 
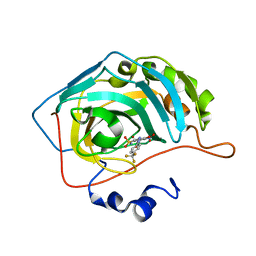 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
3ZWD
 
 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, in complex with the alpha-mannosylglycerate. | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-07-28 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
3ZX5
 
 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, covalently bound to vanadate and in complex with alpha- mannosylglycerate and magnesium | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE, ... | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-08-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
2GRI
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Herrmann, T, Saikatendu, K.S, Joseph, J, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
3DL4
 
 | | Non-Aged Form of Mouse Acetylcholinesterase Inhibited by Tabun- Update | | Descriptor: | Acetylcholinesterase, HEXAETHYLENE GLYCOL | | Authors: | Carletti, E, Li, H, Li, B, Ekstrom, F, Nicolet, Y, Loiodice, M, Gillon, E, Froment, M.T, Lockridge, O, Schopfer, L.M, Masson, P, Nachon, F. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
1SWT
 
 | | CORE-STREPTAVIDIN MUTANT D128A IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Freitag, S, Le Trong, I, Chu, V, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-22 | | Release date: | 1999-07-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural snapshot of an intermediate on the streptavidin-biotin dissociation pathway.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7ZY5
 
 | | Crystal structure of compound 2 bound to CK2alpha | | Descriptor: | 1-NAPHTHOL, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
2JFM
 
 | | CRYSTAL STRUCTURE OF HUMAN STE20-LIKE KINASE (UNLIGANDED FORM) | | Descriptor: | 1,2-ETHANEDIOL, STE20-LIKE SERINE-THREONINE KINASE | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Bunkoczi, G, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-02-02 | | Release date: | 2007-02-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2WU3
 
 | | CRYSTAL STRUCTURE OF MOUSE ACETYLCHOLINESTERASE IN COMPLEX WITH FENAMIPHOS AND HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Hornberg, A, Artursson, E, Warme, R, Pang, Y.-P, Ekstrom, F. | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Oxime-Bound Fenamiphos-Acetylcholinesterases: Reactivation Involving Flipping of the His447 Ring to Form a Reactive Glu334-His447-Oxime Triad.
Biochem.Pharm., 79, 2010
|
|
3ZSY
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (R)-(4-CARBOXY-1,3-BENZODIOXOL-5-YL)METHYL-[[2-(CYCLOHEXYLMETHYLCARBAMOYL)PHENYL]METHYL]-METHYL-AZANIUM, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
8A5G
 
 | |
2JG5
 
 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X08
 
 | | cytochrome c peroxidase: ascorbate bound to the engineered ascorbate binding site | | Descriptor: | ASCORBIC ACID, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
6E2P
 
 | |
1G3N
 
 | |
