5QJ8
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z2856434829 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJA
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1497321453 | | Descriptor: | (2,5-dimethyl-1,3-thiazol-4-yl)(pyrrolidin-1-yl)methanone, 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJT
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z969560582 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QK5
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1267773786 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
3HUK
 
 | | Benzylacetate in complex with T4 lysozyme L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, PHOSPHATE ION, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
5QJQ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z56791867 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4H58
 
 | | BRAF in complex with compound 3 | | Descriptor: | CHLORIDE ION, N-(4-{[(2-methoxyethyl)amino]methyl}phenyl)-6-(pyridin-4-yl)quinazolin-2-amine, Serine/threonine-protein kinase B-raf | | Authors: | Vasbinder, M, Aquila, B, Augustin, M, Chueng, T, Cook, D, Drew, L, Fauber, B, Glossop, S, Godin, R, Grondine, M, Hennessy, E, Johannes, J, Lee, S, Lyne, P, Moertl, M, Omer, C, Palakurthi, S, Pontz, T, Read, J, Sha, L, Shen, M, Steinbacher, S, Wang, H, Wu, A, Ye, M, Bagal, B. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf(V600E) Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
5QK9
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z102895082 | | Descriptor: | 1,2-ETHANEDIOL, 7,8-dimethoxyphthalazin-1(2H)-one, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
7CEI
 
 | | THE ENDONUCLEASE DOMAIN OF COLICIN E7 IN COMPLEX WITH ITS INHIBITOR IM7 PROTEIN | | Descriptor: | PROTEIN (COLICIN E7 IMMUNITY PROTEIN), ZINC ION | | Authors: | Ko, T.P, Liao, C.C, Ku, W.Y, Chak, K.F, Yuan, H.S. | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein.
Structure Fold.Des., 7, 1999
|
|
4H4L
 
 | | Crystal Structure of ternary complex of HutP(HutP-L-His-Zn) | | Descriptor: | HISTIDINE, Hut operon positive regulatory protein, ZINC ION | | Authors: | Dhakshnamoorthy, B, Misono, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2012-09-17 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alternative binding modes of l-histidine guided by metal ions for the activation of the antiterminator protein HutP of Bacillus subtilis.
J.Struct.Biol., 183, 2013
|
|
4E28
 
 | | Structure of human thymidylate synthase in inactive conformation with a novel non-peptidic inhibitor | | Descriptor: | 2-{(2Z,5S)-4-hydroxy-2-[(2E)-(2-hydroxybenzylidene)hydrazinylidene]-2,5-dihydro-1,3-thiazol-5-yl}-N-[3-(trifluoromethyl)phenyl]acetamide, 2-{(5S)-2-[(2E)-2-(2-hydroxybenzylidene)hydrazinyl]-4-oxo-4,5-dihydro-1,3-thiazol-5-yl}-N-[3-(trifluoromethyl)phenyl]acetamide, SULFATE ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M, Costi, M.P. | | Deposit date: | 2012-03-07 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Inhibitor of ovarian cancer cells growth by virtual screening: a new thiazole derivative targeting human thymidylate synthase.
J.Med.Chem., 55, 2012
|
|
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
2FT3
 
 | | Crystal structure of the biglycan dimer core protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Biglycan, CITRATE ANION | | Authors: | Scott, P.G, Dodd, C.M, Bergmann, E.M. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the Biglycan Dimer and Evidence That Dimerization Is Essential for Folding and Stability of Class I Small Leucine-rich Repeat Proteoglycans.
J.Biol.Chem., 281, 2006
|
|
1KKV
 
 | |
1M72
 
 | | Crystal Structure of Caspase-1 from Spodoptera frugiperda | | Descriptor: | 1,2-ETHANEDIOL, Ace-Asp-Glu-Val-Asp-chloromethylketone, Caspase-1 | | Authors: | Forsyth, C.M, Lemongello, D, Friesen, P.D, Fisher, A.J. | | Deposit date: | 2002-07-18 | | Release date: | 2004-01-20 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an invertebrate caspase.
J.Biol.Chem., 279, 2004
|
|
2FVP
 
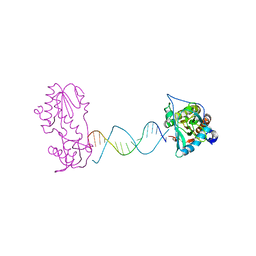 | | A Structural Study of the CA Dinucleotide Step in the Integrase Processing Site of Moloney Murine Leukemia Virus | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*TP*GP*CP*AP*AP*TP*GP*AP*AP*A)-3', Reverse transcriptase | | Authors: | Montano, S.P, Cote, M.L, Roth, M.J, Georgiadis, M.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus.
Nucleic Acids Res., 34, 2006
|
|
1M9N
 
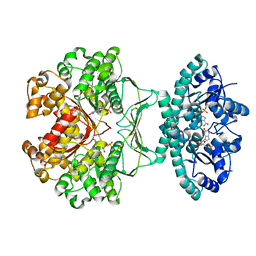 | | CRYSTAL STRUCTURE OF THE HOMODIMERIC BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME AVIAN ATIC IN COMPLEX WITH AICAR AND XMP AT 1.93 ANGSTROMS. | | Descriptor: | AICAR TRANSFORMYLASE-IMP CYCLOHYDROLASE, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Wolan, D.W, Greasly, S.E, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2002-07-29 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Avian AICAR Transformylase Mechanism.
Biochemistry, 41, 2002
|
|
2P82
 
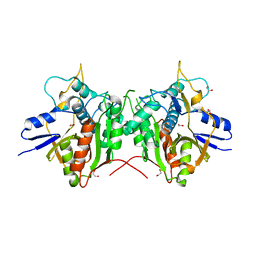 | | Cysteine protease ATG4A | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cysteine protease ATG4A | | Authors: | Walker, J.R, Davis, T, Mujib, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-21 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human cysteine protease ATG4A
To be Published
|
|
4M3H
 
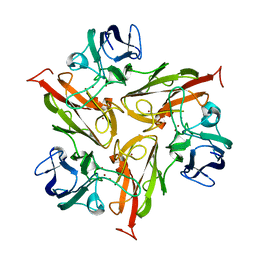 | | Crystal structure of small laccase Ssl1 from Streptomyces sviceus | | Descriptor: | COPPER (II) ION, Copper oxidase, OXYGEN ATOM | | Authors: | Gunne, M, Hoeppner, A, Hagedoorn, P.-L, Urlacher, V.B. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus.
Febs J., 281, 2014
|
|
7LHY
 
 | |
1MDU
 
 | | Crystal structure of the chicken actin trimer complexed with human gelsolin segment 1 (GS-1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Dawson, J.F, Sablin, E.P, Spudich, J.A, Fletterick, R.J. | | Deposit date: | 2002-08-07 | | Release date: | 2003-01-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an F-actin trimer disrupted by gelsolin and implications for the mechanism of severing
J.Biol.Chem., 278, 2003
|
|
7LC9
 
 | |
2YV9
 
 | |
5JSX
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+ and uridine-diphosphate-glucose (UDP-Glc) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
4DK5
 
 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine inhibitor | | Descriptor: | 4-(2-[(6-methoxypyridin-3-yl)amino]-5-{[4-(methylsulfonyl)piperazin-1-yl]methyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-based design of a novel series of potent, selective inhibitors of the class I phosphatidylinositol 3-kinases.
J.Med.Chem., 55, 2012
|
|
