4AMF
 
 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
1JF1
 
 | | Crystal structure of HLA-A2*0201 in complex with a decameric altered peptide ligand from the MART-1/Melan-A | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, ZINC ION, ... | | Authors: | Sliz, P, Michielin, O, Cerottini, J.C, Luescher, I, Romero, P, Karplus, M, Wiley, D.C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes.
J.Immunol., 167, 2001
|
|
5D2J
 
 | |
1OQC
 
 | |
5UPD
 
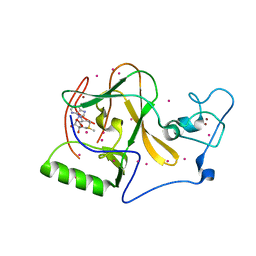 | | Methyltransferase domain of human Wolf-Hirschhorn Syndrome Candidate 1-Like protein 1 (WHSC1L1) | | Descriptor: | Histone-lysine N-methyltransferase NSD3, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION, ... | | Authors: | Tempel, W, Yu, W, Dong, A, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methyltransferase domain of human Wolf-Hirschhorn Syndrome Candidate 1-Like protein 1 (WHSC1L1)
To Be Published
|
|
1ZUU
 
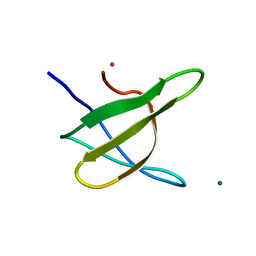 | | Crystal structure of the yeast Bzz1 first SH3 domain at 0.97-A resolution | | Descriptor: | BZZ1 protein, MAGNESIUM ION, UNKNOWN ATOM OR ION | | Authors: | Kursula, P, Kursula, I, Lehmann, F, Zou, P, Song, Y.H, Wilmanns, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structural genomics of yeast SH3 domains
To be Published
|
|
1JHT
 
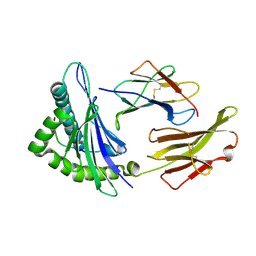 | | Crystal structure of HLA-A2*0201 in complex with a nonameric altered peptide ligand (ALGIGILTV) from the MART-1/Melan-A. | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, beta-2-microglobulin, ... | | Authors: | Sliz, P, Michielin, O, Karplus, M, Romero, P, Wiley, D. | | Deposit date: | 2001-06-28 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes.
J.Immunol., 167, 2001
|
|
2JI2
 
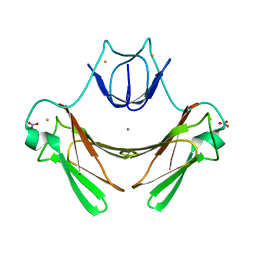 | | X-ray structure of E114A mutant of superoxide reductase from Desulfoarculus baarsii in the native, reduced form | | Descriptor: | CALCIUM ION, Desulfoferrodoxin, FE (II) ION, ... | | Authors: | Katona, G, Carpentier, P, Niviere, V, Amara, P, Adam, V, Ohana, J, Tsanov, N, Bourgeois, D. | | Deposit date: | 2007-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme.
Science, 316, 2007
|
|
3MRW
 
 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution
To be Published
|
|
1TFN
 
 | |
2JLQ
 
 | | Dengue virus 4 NS3 helicase structure, apo enzyme. | | Descriptor: | CHLORIDE ION, GLYCEROL, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
5V00
 
 | | Structure of HutD from Pseudomonas fluorescens SBW25 (Formate condition) | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Liu, Y, Johnston, J.M, Gerth, M.L, Baker, E.N, Lott, J.S, Rainey, P.B. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a bicupin protein HutD involved in histidine utilization in Pseudomonas.
Proteins, 85, 2017
|
|
5D9N
 
 | | Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG | | Descriptor: | B-1,4-endoglucanase, CALCIUM ION, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Morar, M, Stogios, P.J, Xu, X, Cui, H, Di Leo, R, Yim, V, Savchenko, A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Function Analysis of a Mixed-linkage beta-Glucanase/Xyloglucanase from the Key Ruminal Bacteroidetes Prevotella bryantii B14.
J.Biol.Chem., 291, 2016
|
|
2JMG
 
 | | Solution structure of V7R mutant of HIV-1 myristoylated matrix protein | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Loeliger, E, Luncsford, P, Liriano, M, Tai, J, Kim, A, Miller, J, Joshi, A, Freed, E.O, Summers, M.F. | | Deposit date: | 2006-11-11 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the HIV-1 matrix protein turn off the myristyl switch.
J.Mol.Biol., 366, 2007
|
|
2VCD
 
 | | Solution structure of the FKBP-domain of Legionella pneumophila Mip in complex with rapamycin | | Descriptor: | Outer membrane protein MIP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Paschke, A.-K, Fischer, G, Roesch, P, Faber, C. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Legionella pneumophila Mip-rapamycin complex.
BMC Struct. Biol., 8, 2008
|
|
1YFC
 
 | | Solution nmr structure of a yeast iso-1-ferrocytochrome C | | Descriptor: | HEME C, YEAST ISO-1-FERROCYTOCHROME C | | Authors: | Baistrocchi, P, Banci, L, Bertini, I, Turano, P, Bren, K.L, Gray, H.B. | | Deposit date: | 1996-08-08 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Saccharomyces cerevisiae reduced iso-1-cytochrome c.
Biochemistry, 35, 1996
|
|
1YS7
 
 | | Crystal structure of the response regulator protein prrA complexed with Mg2+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The structural basis of signal transduction for the response regulator PrrA from Mycobacterium tuberculosis.
J.Biol.Chem., 281, 2006
|
|
5D3Z
 
 | | Crystal structure of the thioesterase domain of deoxyerythronolide B synthase in complex with a small phosphonate inhibitor | | Descriptor: | CALCIUM ION, Erythronolide synthase, modules 5 and 6, ... | | Authors: | Bergeret, F, Argyropoulos, P, Boddy, C.N, Schmeing, T.M. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards a characterization of the structural determinants of specificity in the macrocyclizing thioesterase for deoxyerythronolide B biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
1PZR
 
 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS2 and DEBS3: the B domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
1G80
 
 | | NMR SOLUTION STRUCTURE OF D(GCGTACGC)2 | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
3MM5
 
 | | Dissimilatory sulfite reductase in complex with the substrate sulfite | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
1JW1
 
 | |
1G1K
 
 | | COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDING PROTEIN | | Authors: | Spinelli, S, Fierobe, H.-P, Belaich, A, Belaich, J.-P, Henrissat, B, Cambillau, C. | | Deposit date: | 2000-10-12 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cohesin module from Clostridium cellulolyticum: implications for dockerin recognition.
J.Mol.Biol., 304, 2000
|
|
3WNQ
 
 | | Crystal structure of (R)-carbonyl reductase H49A mutant from Candida Parapsilosis in complex with 2-hydroxyacetophenone | | Descriptor: | (R)-specific carbonyl reductase, 2-hydroxy-1-phenylethanone, ZINC ION | | Authors: | Wang, S.S, Nie, Y, Xu, Y, Zhang, R.Z, Huang, C.H, Chan, H.C, Guo, R.T, Ko, T.P, Xiao, R. | | Deposit date: | 2013-12-15 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Unconserved substrate-binding sites direct the stereoselectivity of medium-chain alcohol dehydrogenase
Chem.Commun.(Camb.), 50, 2014
|
|
1PZQ
 
 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS 2 and DEBS 3: The A domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
