7SNS
 
 | | 1.55A Resolution Structure of NanoLuc Luciferase | | 分子名称: | ACETATE ION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | 著者 | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | 登録日 | 2021-10-28 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | 1.55A Resolution Structure of NanoLuc Luciferase
To be published
|
|
6MU6
 
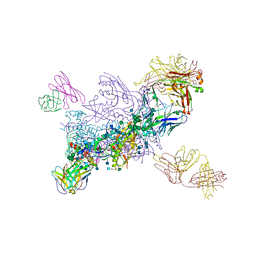 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-814508 in Complex with Human Antibodies 3H109L and 35O22 at 3.2 Angstrom | | 分子名称: | (2R)-{1-[{7-[2-({[3-(dimethylamino)propyl](methyl)amino}methyl)-1,3-thiazol-4-yl]-4-methoxy-1H-pyrrolo[2,3-c]pyridin-3-yl}(oxo)acetyl]piperidin-4-yl}(phenyl)acetonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lai, Y.-T, Kwong, P.D. | | 登録日 | 2018-10-22 | | 公開日 | 2019-01-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.551 Å) | | 主引用文献 | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
7ZHR
 
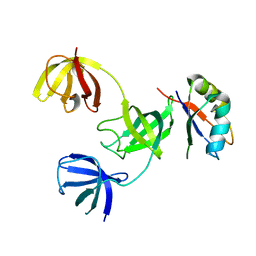 | |
5K2B
 
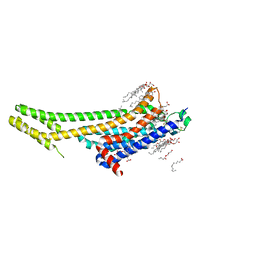 | | 2.5 angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | 著者 | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | 登録日 | 2016-05-18 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
3D0X
 
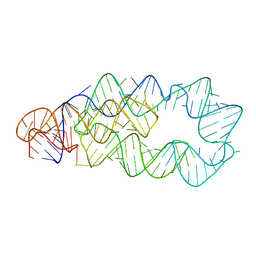 | |
8F2B
 
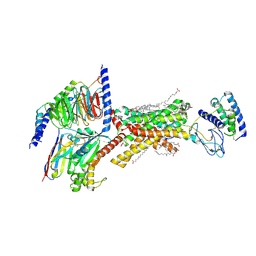 | | Amylin 3 Receptor in complex with Gs and Pramlintide analogue peptide San45 | | 分子名称: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Cao, J, Sexton, P.M, Wootten, D.L, Belousoff, M.J. | | 登録日 | 2022-11-07 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2 Å) | | 主引用文献 | Structural insight into selectivity of amylin and calcitonin receptor agonists.
Nat.Chem.Biol., 20, 2024
|
|
4R8Y
 
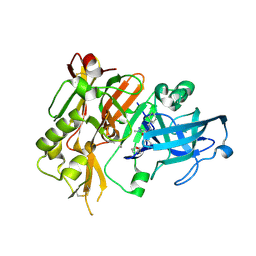 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((R)-1-(2-cyclopentylacetyl)pyrrolidin-3-yl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | 分子名称: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(3R)-1-(cyclopentylacetyl)pyrrolidin-3-yl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Caldwell, J.P, Strickland, C. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4QT4
 
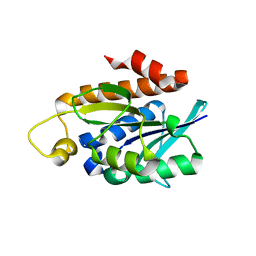 | | Crystal structure of Peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 Angstrom resolution shows the Closed Structure of the Substrate Binding Cleft | | 分子名称: | Peptidyl-tRNA hydrolase | | 著者 | Singh, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2014-07-07 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 angstrom resolution shows the closed structure of the substrate-binding cleft.
FEBS Open Bio, 4, 2014
|
|
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | 登録日 | 2022-05-20 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5K6C
 
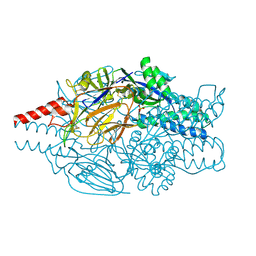 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 variant. | | 分子名称: | Fusion glycoprotein F0,Fusion glycoprotein F0, SULFATE ION | | 著者 | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | 登録日 | 2016-05-24 | | 公開日 | 2016-10-12 | | 最終更新日 | 2016-10-19 | | 実験手法 | X-RAY DIFFRACTION (3.576 Å) | | 主引用文献 | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6N0Y
 
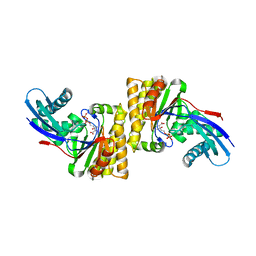 | |
5KE3
 
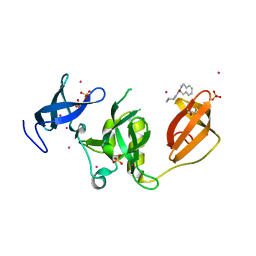 | | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a | | 分子名称: | (S)-N-(furan-2-ylmethyl)-1-(1,2,3,4-tetrahydroisoquinoline-3-carbonyl)piperidine-4-carboxamide, BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETDB1, ... | | 著者 | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Ferreira de Freitas, R, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2016-06-09 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a
to be published
|
|
6YEW
 
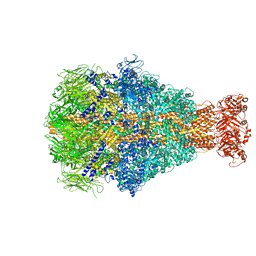 | | Morganella morganii TcdA4 in complex with porcine mucosa heparin | | 分子名称: | Insecticidal toxin protein | | 著者 | Roderer, D, Broecker, F, Sitsel, O, Kaplonek, P, Leidreiter, F, Seeberger, P.H, Raunser, S. | | 登録日 | 2020-03-25 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Glycan-dependent cell adhesion mechanism of Tc toxins.
Nat Commun, 11, 2020
|
|
6PX0
 
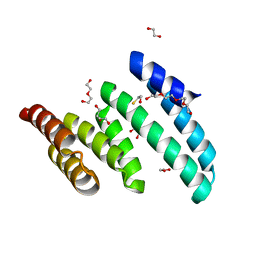 | |
6G72
 
 | | Mouse mitochondrial complex I in the deactive state | | 分子名称: | ACETYL GROUP, ADENOSINE-5'-DIPHOSPHATE, Acyl carrier protein, ... | | 著者 | Agip, A.N.A, Blaza, J.N, Bridges, H.R, Viscomi, C, Rawson, S, Muench, S.P, Hirst, J. | | 登録日 | 2018-04-04 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6MX2
 
 | |
3CG1
 
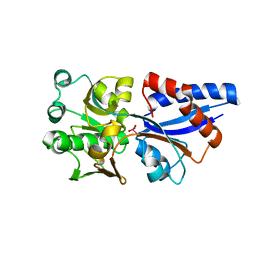 | |
6Q2I
 
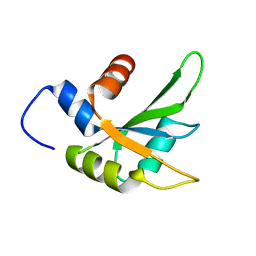 | |
8Q3X
 
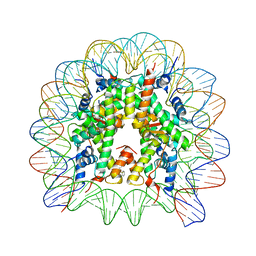 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Kaposi Sarcoma Associated Herpesvirus LANA Peptide-Au[I] Compound) | | 分子名称: | 4-diphenylphosphanylbenzoic acid, DNA (145-MER), GOLD ION, ... | | 著者 | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | 登録日 | 2023-08-04 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
6G0O
 
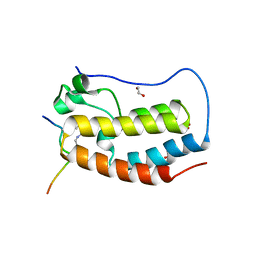 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated ATRX peptide (K1030ac/K1033ac) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcriptional regulator ATRX | | 著者 | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | 登録日 | 2018-03-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | 著者 | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | 登録日 | 2016-05-18 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5MZK
 
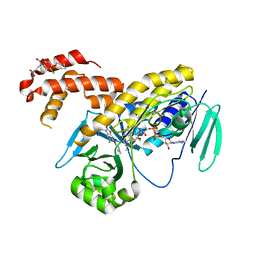 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with 3-[5-chloro-6-(cyclobutylmethoxy)-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl]propanoic acid | | 分子名称: | 3-[5-chloro-6-(cyclobutylmethoxy)-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl]propanoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Rowland, P. | | 登録日 | 2017-01-31 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Development of a Series of Kynurenine 3-Monooxygenase Inhibitors Leading to a Clinical Candidate for the Treatment of Acute Pancreatitis.
J. Med. Chem., 60, 2017
|
|
6YEC
 
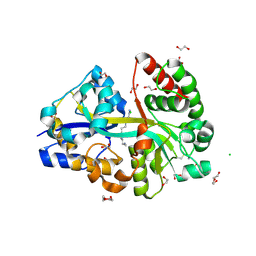 | | E.coli's Putrescine receptor PotF complexed with Spermine | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | 登録日 | 2020-03-24 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
4R9I
 
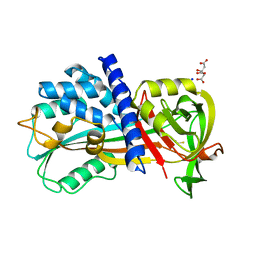 | | Crystal structure of cysteine proteinase inhibitor Serpin18 from Bombyx mori | | 分子名称: | BETA-MERCAPTOETHANOL, CITRATE ANION, SODIUM ION, ... | | 著者 | Guo, P.C, He, H.W, Zhao, P, Xia, Q.Y. | | 登録日 | 2014-09-05 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural insights into the unique inhibitory mechanism of the silkworm protease inhibitor serpin18
Sci Rep, 5, 2015
|
|
4QX1
 
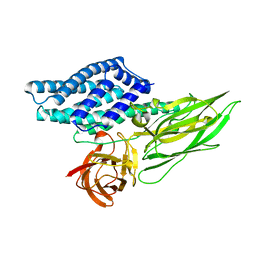 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | 分子名称: | Pesticidal crystal protein cry3Aa | | 著者 | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | 登録日 | 2014-07-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
