1I9N
 
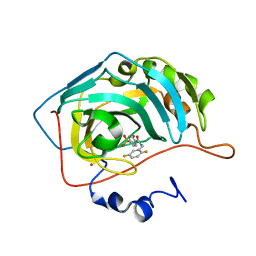 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,5-DIFLUOROPHENYL)METHYL]-BENZAMIDE | | 分子名称: | 4-(AMINOSULFONYL)-N-[(2,5-DIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | 著者 | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | 登録日 | 2001-03-20 | | 公開日 | 2001-03-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
3M8W
 
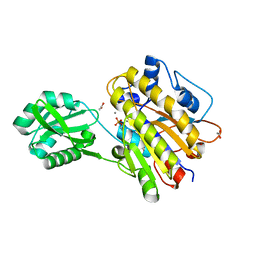 | | Phosphopentomutase from Bacillus cereus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, MANGANESE (II) ION, ... | | 著者 | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2010-03-19 | | 公開日 | 2010-12-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
2YOX
 
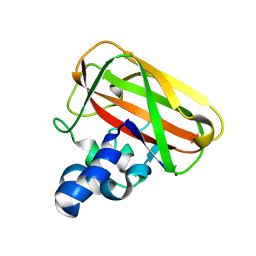 | | Bacillus amyloliquefaciens CBM33 in complex with Cu(I) after photoreduction | | 分子名称: | COPPER (I) ION, RBAM17540 | | 著者 | Hemsworth, G.R, Taylor, E.J, Kim, R.Q, Lewis, S.J, Turkenburg, J.P, Davies, G.J, Walton, P.H. | | 登録日 | 2012-10-29 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Copper Active Site of Cbm33 Polysaccharide Oxygenases.
J.Am.Chem.Soc., 135, 2013
|
|
1APJ
 
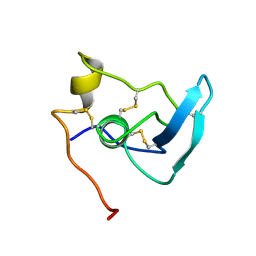 | | NMR STUDY OF THE TRANSFORMING GROWTH FACTOR BETA BINDING PROTEIN-LIKE DOMAIN (TB MODULE/8-CYS DOMAIN), NMR, 21 STRUCTURES | | 分子名称: | FIBRILLIN | | 著者 | Yuan, X, Downing, A.K, Knott, V, Handford, P.A. | | 登録日 | 1997-07-22 | | 公開日 | 1998-01-28 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the transforming growth factor beta-binding protein-like module, a domain associated with matrix fibrils.
EMBO J., 16, 1997
|
|
1FL8
 
 | |
5AG9
 
 | | CRYSTAL STRUCTURE OF A MUTANT (665sXa) C-TERMINAL DOMAIN OF RGPB | | 分子名称: | Gingipain R2, SULFATE ION | | 著者 | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | 登録日 | 2015-01-29 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain.
Sci Rep, 6, 2016
|
|
6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | 登録日 | 2020-03-03 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | COVID-19 main protease with unliganded active site
To Be Published
|
|
3CIJ
 
 | |
1B4L
 
 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | 分子名称: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | 著者 | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | 登録日 | 1998-12-22 | | 公開日 | 1999-12-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
2WDU
 
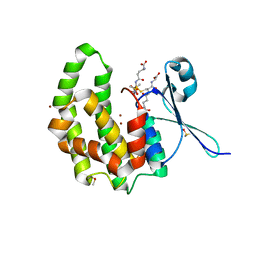 | | Fasciola hepatica sigma class GST | | 分子名称: | BROMIDE ION, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | 著者 | Line, K, Isupov, M.N, LaCourse, E.J, Brophy, P.M, Littlechild, J.A. | | 登録日 | 2009-03-26 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | The 1.6 Angstrom Crystal Structure of the Fasciola Hepatica Sigma Class Gst
To be Published
|
|
1W60
 
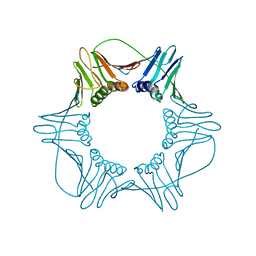 | | NATIVE HUMAN PCNA | | 分子名称: | PROLIFERATING CELL NUCLEAR ANTIGEN | | 著者 | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M. | | 登録日 | 2004-08-11 | | 公開日 | 2005-01-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2WC4
 
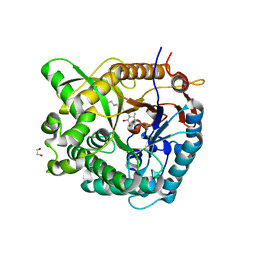 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-thia-(+)-castanospermine | | 分子名称: | (3Z,5S,6R,7S,8R,8aS)-3-(octylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | 登録日 | 2009-03-08 | | 公開日 | 2009-04-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
4E8O
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ih from Acinetobacter baumannii | | 分子名称: | Aac(6')-Ih protein, CHLORIDE ION | | 著者 | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-03-20 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.138 Å) | | 主引用文献 | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
1BA4
 
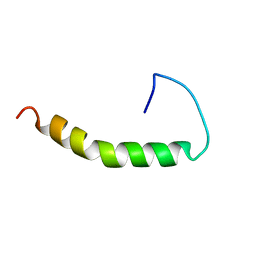 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | 分子名称: | AMYLOID BETA-PEPTIDE | | 著者 | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | 登録日 | 1998-04-07 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
5ML9
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | 著者 | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | 登録日 | 2016-12-06 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2WD3
 
 | | Highly Potent First Examples of Dual Aromatase-Steroid Sulfatase Inhibitors based on a Biphenyl Template | | 分子名称: | 3-CHLORO-2'-CYANO-5'-(1H-1,2,4-TRIAZOL-1-YLMETHYL)BIPHENYL-4-YL SULFAMATE, CARBONIC ANHYDRASE 2, ZINC ION | | 著者 | Woo, L.W.L, Jackson, T, Putey, A, Cozier, G, Leonard, P, Acharya, K.R, Chander, S.K, Purohit, A, Reed, M.J, Potter, B.V.L. | | 登録日 | 2009-03-19 | | 公開日 | 2010-02-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Highly Potent First Examples of Dual Aromatase-Steroid Sulfatase Inhibitors Based on a Biphenyl Template.
J.Med.Chem., 53, 2010
|
|
6YCE
 
 | | Structure the bromelain protease from Ananas comosus with a thiomethylated active cysteine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FBSB, ISOPROPYL ALCOHOL, ... | | 著者 | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | 登録日 | 2020-03-18 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
2W7E
 
 | | Crystal structure of Y51FbsSHMT obtained in the presence of Glycine | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, PHOSPHATE ION, ... | | 著者 | Rajaram, V, Bhavani, B.S, Bisht, S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | 登録日 | 2008-12-22 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Importance of Tyrosine Residues of Bacillus Stearothermophilus Serine Hydroxymethyltransferase in Cofactor Binding and L-Allo-Thr Cleavage.
FEBS J., 275, 2008
|
|
5AAE
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14d) | | 分子名称: | 3-((4-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)-1H-pyrazol-1-yl)methyl)-5-methylisoxazole, AURORA KINASE A | | 著者 | McIntyre, P.J, Bayliss, R. | | 登録日 | 2015-07-24 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3ZHV
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, post-decarboxylation intermediate from pyruvate (2-hydroxyethyl-ThDP) | | 分子名称: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-[(1S)-1-oxidanylethyl]-1,3-thiazol-3-ium-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | 登録日 | 2012-12-24 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
2WB4
 
 | | activated diguanylate cyclase PleD in complex with c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), BERYLLIUM TRIFLUORIDE ION, DIGUANYLATE CYCLASE, ... | | 著者 | Wassmann, P, Schirmer, T. | | 登録日 | 2009-02-20 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of Activated Pled, Identification of Dimerization and Catalysis Relevant Regulatory Mechanisms
To be Published
|
|
5LVC
 
 | | Aichi virus 1: empty particle | | 分子名称: | VP0, VP1, VP3 | | 著者 | Sabin, C, Fuzik, T, Skubnik, K, Palkova, L, Lindberg, A.M, Plevka, P. | | 登録日 | 2016-09-13 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structure of Aichi Virus 1 and Its Empty Particle: Clues to Kobuvirus Genome Release Mechanism.
J.Virol., 90, 2016
|
|
5LEL
 
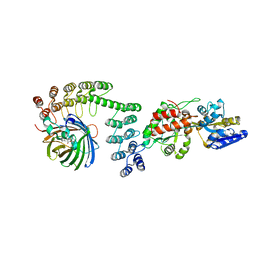 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | 分子名称: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-30 | | 公開日 | 2017-11-15 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
1I3Q
 
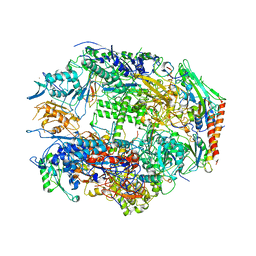 | | RNA POLYMERASE II CRYSTAL FORM I AT 3.1 A RESOLUTION | | 分子名称: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | 著者 | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | 登録日 | 2001-02-15 | | 公開日 | 2001-04-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
2WGW
 
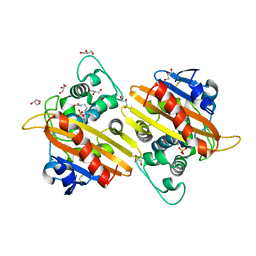 | | Crystal structure of the OXA-10 V117T mutant at pH 8.0 | | 分子名称: | BETA-LACTAMASE OXA-10, GLYCEROL, SULFATE ION | | 著者 | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | 登録日 | 2009-04-27 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
