3MVD
 
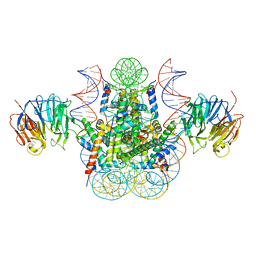 | | Crystal structure of the chromatin factor RCC1 in complex with the nucleosome core particle | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Makde, R.D, England, J.R, Yennawar, H.P, Tan, S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of RCC1 chromatin factor bound to the nucleosome core particle.
Nature, 467, 2010
|
|
8C75
 
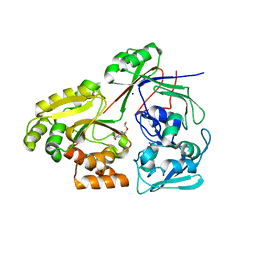 | | PBP AccA from A. tumefaciens Bo542 in apoform 3 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
4RU5
 
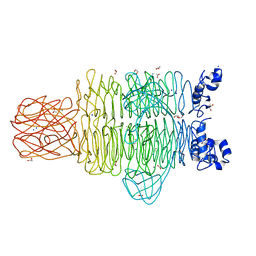 | | Crystal Structure of the Pseudomonas phage phi297 tailspike gp61 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Browning, C, Sycheva, L.V, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2014-11-18 | | Release date: | 2015-11-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence.
Sci Rep, 7, 2017
|
|
6N4R
 
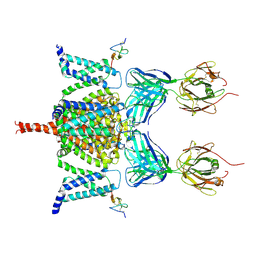 | | CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
6TV7
 
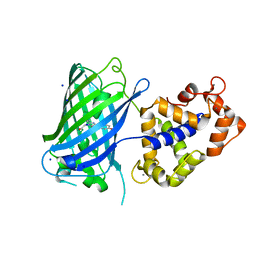 | | Crystal structure of rsGCaMP in the OFF state (illuminated) | | Descriptor: | CALCIUM ION, SODIUM ION, rsGCaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-01-09 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
4RZ8
 
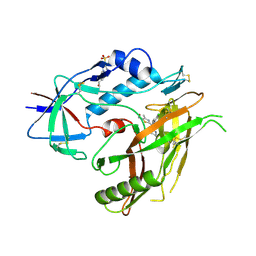 | | Crystal structure of HIV-1 gp120 core in complex with NBD-11021, a small molecule CD4-antagonist | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-chlorophenyl)-N-{(S)-[5-(hydroxymethyl)-4-methyl-1,3-thiazol-2-yl][(2R)-piperidin-2-yl]methyl}-1H-pyrrole-2-carboxamide, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of a Small Molecule CD4-Antagonist with Broad Spectrum Anti-HIV-1 Activity.
J.Med.Chem., 58, 2015
|
|
8C2Z
 
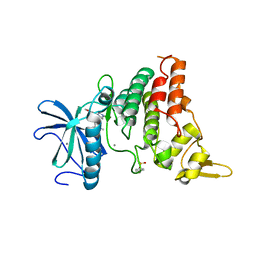 | | Crystal structure of DYRK1B in complex with AZ191 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1B, MANGANESE (II) ION, N-[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]-4-(1-methylpyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural perspective on the design of selective DYRK1B inhibitors
To Be Published
|
|
6E3Y
 
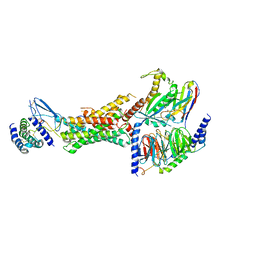 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
8C6W
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 1 | | Descriptor: | Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8C5G
 
 | |
6MZJ
 
 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 2 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6N1Q
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in D2 symmetry | | Descriptor: | DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6N1B
 
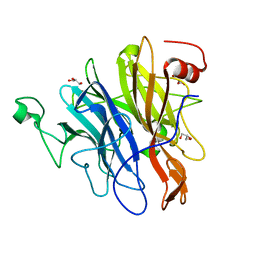 | | Crystal structure of an N-acetylgalactosamine deacetylase from F. plautii in complex with blood group B trisaccharide | | Descriptor: | CALCIUM ION, Carbohydrate-binding protein, GLYCEROL, ... | | Authors: | Sim, L, Rahfeld, P, Withers, S.G. | | Deposit date: | 2018-11-08 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An enzymatic pathway in the human gut microbiome that converts A to universal O type blood.
Nat Microbiol, 4, 2019
|
|
8U50
 
 | |
6MW0
 
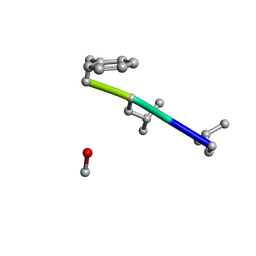 | | Mle-Phe-Mle-D-Phe. Linear tetrapeptide related to pseudoxylallemycin A. | | Descriptor: | METHANOL, Mle-Phe-Mle-D-Phe Linear tetrapeptide related to pseudoxylallemycin A | | Authors: | Cameron, A.J, Harris, P.W.R, Brimble, M.A, Squire, C.J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Investigations of the key macrolactamisation step in the synthesis of cyclic tetrapeptide pseudoxylallemycin A.
Org.Biomol.Chem., 17, 2019
|
|
6JJ9
 
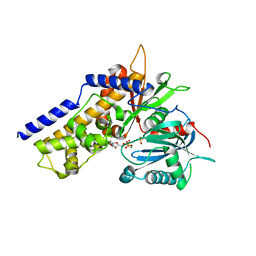 | | Crystal structure of OsHXK6-Glc-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hexokinase-6, MAGNESIUM ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of OsHXK6-Glc-ATP-Mg2+ complex
To Be Published
|
|
8U51
 
 | |
6WCQ
 
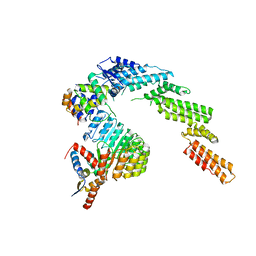 | | Structure of a substrate-bound DQC ubiquitin ligase | | Descriptor: | Cullin-1, F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, ... | | Authors: | Mena, E.L, Jevtic, P, Greber, B.J, Gee, C.L, Lew, B.G, Akopian, D, Nogales, E, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
2HD7
 
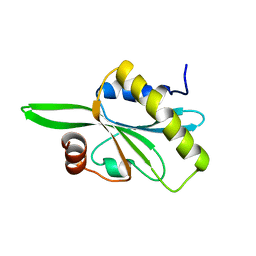 | | Solution structure of C-teminal domain of twinfilin-1. | | Descriptor: | Twinfilin-1 | | Authors: | Hellman, M.H, Paavilainen, V.O, Annila, A, Lappalainen, P, Permi, P.I. | | Deposit date: | 2006-06-20 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis and evolutionary origin of actin filament capping by twinfilin
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3JTL
 
 | |
3R2R
 
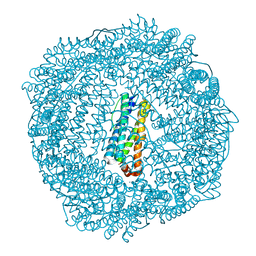 | | 1.65A resolution structure of Iron Soaked FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION, ... | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
8V52
 
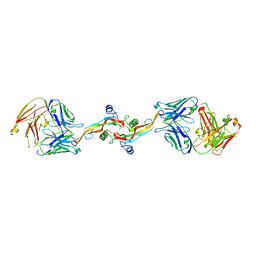 | |
3JW6
 
 | |
3ONY
 
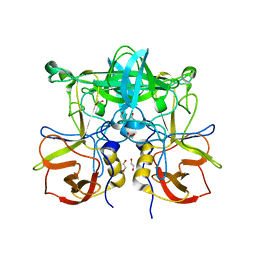 | | Crystal Structure of P Domain from Norwalk Virus Strain Vietnam 026 in complex with Fucose | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-08-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of GII.10 and GII.12 Norovirus Protruding Domains in Complex with Histo-Blood Group Antigens Reveal Details for a Potential Site of Vulnerability.
J.Virol., 85, 2011
|
|
4TKI
 
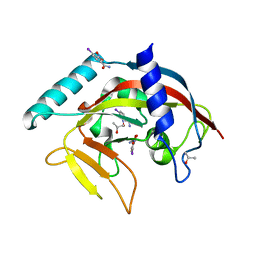 | | Crystal Structure of human Tankyrase 2 in complex with BSI-201. | | Descriptor: | 4-iodo-3-nitrobenzamide, ISOPROPYL ALCOHOL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
