1MOM
 
 | | CRYSTAL STRUCTURE OF MOMORDIN, A TYPE I RIBOSOME INACTIVATING PROTEIN FROM THE SEEDS OF MOMORDICA CHARANTIA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MOMORDIN | | Authors: | Husain, J, Tickle, I.J, Wood, S.P. | | Deposit date: | 1994-03-04 | | Release date: | 1994-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of momordin, a type I ribosome inactivating protein from the seeds of Momordica charantia.
FEBS Lett., 342, 1994
|
|
8DWZ
 
 | | CA domain of VanSA histidine kinase, 7 keV data | | Descriptor: | CADMIUM ION, Sensor protein VanS | | Authors: | Loll, P.J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of VanS from vancomycin-resistant enterococci: A sensor kinase with weak ATP binding.
J.Biol.Chem., 299, 2023
|
|
2FYT
 
 | | Human HMT1 hnRNP methyltransferase-like 3 (S. cerevisiae) protein | | Descriptor: | Protein arginine N-methyltransferase 3, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-08 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP
methyltransferase-like 3 in complex with SAH.
To be Published
|
|
2G2Q
 
 | | The crystal structure of G4, the poxviral disulfide oxidoreductase essential for cytoplasmic disulfide bond formation | | Descriptor: | Glutaredoxin-2, SULFATE ION | | Authors: | Su, H.P, Lin, D.Y, Garboczi, D.N. | | Deposit date: | 2006-02-16 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of G4, the poxvirus disulfide oxidoreductase essential for virus maturation and infectivity.
J.Virol., 80, 2006
|
|
7YG4
 
 | | Structure of WTAP-VIRMA in the m6A writer complex | | Descriptor: | Pre-mRNA-splicing regulator WTAP, Protein virilizer homolog | | Authors: | Yan, X.H, Guan, Z.Y, Tang, C, Yin, P. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AI-empowered integrative structural characterization of m 6 A methyltransferase complex.
Cell Res., 32, 2022
|
|
7Y75
 
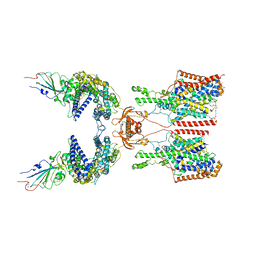 | | SIT1-ACE2-BA.2 RBD | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, Y.P, Li, Y.N, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-21 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2.
Cell Discov, 8, 2022
|
|
2FZK
 
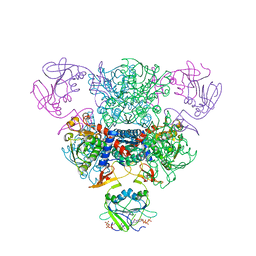 | | The Structure of Wild-Type E. Coli Aspartate Transcarbamoylase in Complex with Novel T State Inhibitors at 2.50 Resolution | | Descriptor: | 3,5-BIS[(PHOSPHONOACETYL)AMINO]BENZOIC ACID, Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Heng, S, Stieglitz, K.A, Eldo, J, Xia, J, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2006-02-09 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T-state Inhibitors of E. coli Aspartate Transcarbamoylase that Prevent the Allosteric Transition.
Biochemistry, 45, 2006
|
|
2G0S
 
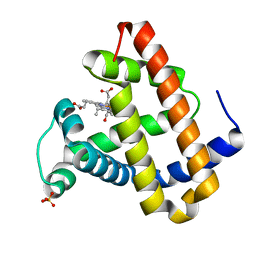 | | Unphotolyzed CO-bound L29F Myoglobin, crystal 2 | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Aranda, R, Levin, E.J, Schotte, F, Anfinrud, P.A, Phillips Jr, G.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2G0Z
 
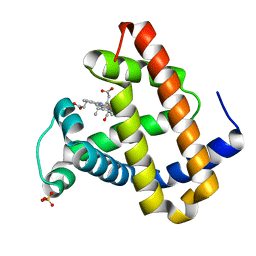 | | Photolyzed CO L29F Myoglobin: 1ns | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Aranda, R, Levin, E.J, Schotte, F, Anfinrud, P.A, Phillips Jr, G.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2G6E
 
 | | Structure of cyclized F64L S65A Y66S GFP variant | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P. | | Deposit date: | 2006-02-24 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Understanding GFP Posttranslational Chemistry: Structures of Designed Variants that Achieve Backbone Fragmentation, Hydrolysis, and Decarboxylation.
J.Am.Chem.Soc., 128, 2006
|
|
6KJ5
 
 | | Crystal structure of 10-Hydroxygeraniol Dehydrogenase apo form from Cantharanthus roseus | | Descriptor: | 10-hydroxygeraniol dehydrogenase, ZINC ION | | Authors: | Sandholu, A.S, Sharmila, P.M, Thulasiram, H.V, Kulkarni, K.A. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structural studies on 10-hydroxygeraniol dehydrogenase: A novel linear substrate-specific dehydrogenase from Catharanthus roseus.
Proteins, 88, 2020
|
|
2G2S
 
 | |
6KK2
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 2 | | Descriptor: | 1-[(10~{S},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.2.2]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-07-23 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
2GAF
 
 | | Crystal Structure of the Vaccinia Polyadenylate Polymerase Heterodimer (apo form) | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit | | Authors: | Moure, C.M, Bowman, B.R, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2006-03-08 | | Release date: | 2006-05-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity.
Mol.Cell, 22, 2006
|
|
6KML
 
 | | 2.09 Angstrom resolution crystal structure of tetrameric HigBA toxin-antitoxin complex from E.coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
2G8E
 
 | | Calpain 1 proteolytic core in complex with SNJ-1715, a cyclic hemiacetal-type inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Calpain-1 catalytic subunit, ... | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
6KMQ
 
 | | 2.3 Angstrom resolution structure of dimeric HigBA toxin-antitoxin complex from E. coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
2GHD
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
6KM9
 
 | | Crystal structure of SucA from Vibrio vulnificus | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Seo, P.W, Kim, J.S. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Catalysis Science And Technology, 2020
|
|
7Y5N
 
 | | Structure of 1:1 PAPP-A.ProMBP complex(half map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone marrow proteoglycan, ... | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
4FNQ
 
 | | Crystal structure of GH36 alpha-galactosidase AgaB from Geobacillus stearothermophilus | | Descriptor: | 1,2-ETHANEDIOL, Alpha-galactosidase AgaB | | Authors: | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
3HFO
 
 | |
3UU4
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in the crystal in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7Y76
 
 | | SIT1-ACE2-BA.5 RBD | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, Y.P, Li, Y.N, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-21 | | Release date: | 2023-01-04 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2.
Cell Discov, 8, 2022
|
|
2GF2
 
 | | Crystal structure of human hydroxyisobutyrate dehydrogenase | | Descriptor: | 3-hydroxyisobutyrate dehydrogenase | | Authors: | Papagrigoriou, E, Salah, E, Turnbull, A.P, Smee, C, Burgess, N, Gileadi, O, von Delft, F, Gorrec, F, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human hydroxyisobutyrate dehydrogenase
To be Published
|
|
