4LBV
 
 | | Identifying ligand binding hot spots in proteins using brominated fragments | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Elongation factor Tu-A, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5PJ1
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 50) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PJF
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 64) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PJV
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 80) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2VB3
 
 | | Crystal structure of Ag(I)CusF | | Descriptor: | CATION EFFLUX SYSTEM PROTEIN CUSF, SILVER ION | | Authors: | Xue, Y, Davis, A.V, Balakrishnan, G, Stasser, J.P, Staehlin, B.M, Focia, P, Spiro, T.G, Penner-Hahn, J.E, O'Halloran, T.V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Cu(I) Recognition Via Cation-Pi and Methionine Interactions in Cusf.
Nat.Chem.Biol., 4, 2008
|
|
5PKB
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 96) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PKQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 111) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PL6
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 127) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PLV
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 152) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6G79
 
 | | Coupling specificity of heterotrimeric Go to the serotonin 5-HT1B receptor | | Descriptor: | 2-[5-[2-[4-(4-cyanophenyl)piperazin-1-yl]-2-oxidanylidene-ethoxy]-1~{H}-indol-3-yl]ethylazanium, 5-hydroxytryptamine receptor 1B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Garcia-Nafria, J, Nehme, R, Edwards, P, Tate, C.G. | | Deposit date: | 2018-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM structure of the serotonin 5-HT1Breceptor coupled to heterotrimeric Go.
Nature, 558, 2018
|
|
1P3F
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
5PM3
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 161) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PHB
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09447a | | Descriptor: | 1,2-ETHANEDIOL, 6-azanyl-3,4-dihydro-2~{H}-naphthalen-1-one, Lysine-specific demethylase 4D, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.338 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PMH
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 175) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7XPE
 
 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 8.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
5PHT
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 6) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PMZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 193) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PI9
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 22) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3P9I
 
 | | Crystal structure of perennial ryegrass LpOMT1 complexed with S-adenosyl-L-homocysteine and sinapaldehyde | | Descriptor: | (2E)-3-(4-hydroxy-3,5-dimethoxyphenyl)prop-2-enal, BETA-MERCAPTOETHANOL, Caffeic acid O-methyltransferase, ... | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2010-10-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference.
Plant Cell, 22, 2010
|
|
5PIR
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 40) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5AN7
 
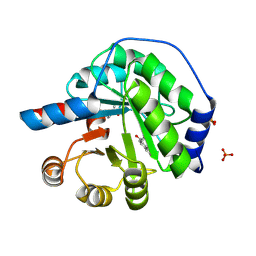 | | Structure of the engineered retro-aldolase RA95.5-8F with a bound 1,3-diketone inhibitor | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, PHOSPHATE ION, RA95.5-8F | | Authors: | Obexer, R, Mittl, P.R.E, Hilvert, D. | | Deposit date: | 2015-09-04 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
5PN7
 
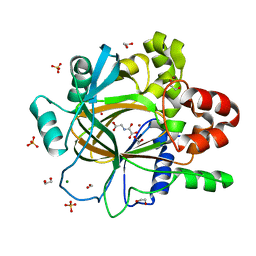 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 201) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2BOY
 
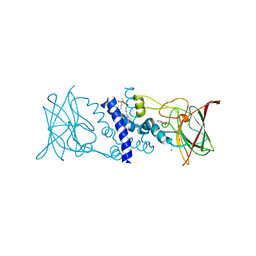 | | Crystal structure of 3-ChloroCatechol 1,2-Dioxygenase from Rhodococcus Opacus 1CP | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 3-CHLOROCATECHOL 1,2-DIOXYGENASE, BENZHYDROXAMIC ACID, ... | | Authors: | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | Deposit date: | 2005-04-15 | | Release date: | 2006-08-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 3-chlorocatechol 1,2-dioxygenase key enzyme of a new modified ortho-pathway from the Gram-positive Rhodococcus opacus 1CP grown on 2-chlorophenol.
J. Mol. Biol., 360, 2006
|
|
4EOF
 
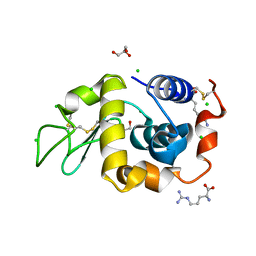 | | Lysozyme in the presence of arginine | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-04-14 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation.
Sci Rep, 6, 2016
|
|
5PJ7
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 56) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
