6QE0
 
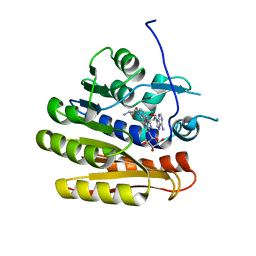 | | Structure of E.coli RlmJ in complex with a bisubstrate analogue (BA2) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[2-[[9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]ethyl]amino]-2-azanyl-butanoic acid, Ribosomal RNA large subunit methyltransferase J | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-03 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.394 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
4PBU
 
 | | Serial Time-resolved crystallography of Photosystem II using a femtosecond X-ray laser The S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R.A, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
5DUJ
 
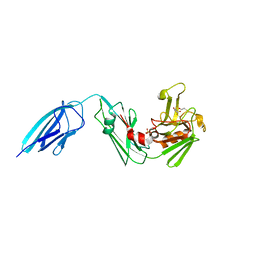 | | Crystal structure of ldtMt2 in complex with Faropenem adduct | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, BETA-MERCAPTOETHANOL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
3NAN
 
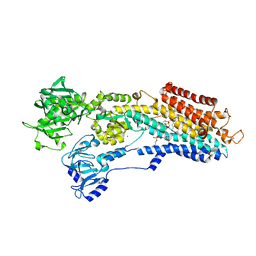 | | SR Ca(2+)-ATPase in the HnE2 state complexed with a Thapsigargin derivative Boc-(phi)Tg | | Descriptor: | (3S,3aR,4S,6S,6aR,7S,8S,9R,9aS,9bS)-6-(acetyloxy)-4-{[4-(3-{6-[(tert-butoxycarbonyl)amino]hexyl}-4-hydroxyphenyl)butanoyl]oxy}-3,3a-dihydroxy-3,6,9-trimethyl-8-{[(2Z)-2-methylbut-2-enoyl]oxy}-2-oxododecahydroazuleno[4,5-b]furan-7-yl octanoate, MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Winther, A.M.L, Sonntag, Y, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs
J.Biol.Chem., 285, 2010
|
|
1Y84
 
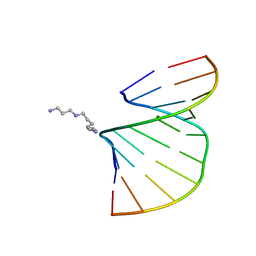 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-[2-(imidazolyl)ethyl] Thymidine (T*) | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(EIT)P*AP*CP*GP*C)-3', MAGNESIUM ION, SPERMINE | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | Deposit date: | 2004-12-10 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
6QHT
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 376 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
3NDA
 
 | | Crystal structure of serpin from tick Ixodes ricinus | | Descriptor: | PENTAETHYLENE GLYCOL, Serpin-2 | | Authors: | Rezacova, P, Kovarova, Z, Chmelar, J, Mares, M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation.
Blood, 117, 2011
|
|
6QI2
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 13536 ms | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6H10
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR073 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.104 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand NR073
To be published
|
|
6QHZ
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 6788 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QKW
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Fluoroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
7LQD
 
 | | Structure of Human MPS1 (TTK) covalently bound to RMS-07 inhibitor | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Dual specificity protein kinase TTK or monopolar spindle 1 kinase, GLYCEROL, ... | | Authors: | Santiago, A.S, dos Reis, C.V, Serafim, R.A.M, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-02-13 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of the First Covalent Monopolar Spindle Kinase 1 (MPS1/TTK) Inhibitor.
J.Med.Chem., 65, 2022
|
|
3R55
 
 | | Crystal structure of the complex of goat lactoperoxidase with Pyrazinamide at 2.1 A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, R.P, Pandey, N, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with Pyrazinamide at 2.1 A resolution
To be Published
|
|
6CM2
 
 | | SAMHD1 HD domain bound to decitabine triphosphate | | Descriptor: | 6-amino-3-{2-deoxy-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-3,4-dihydro-1,3,5-triazin-2(1H)-one, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Oellerich, T, Schneider, C, Thomas, D, Knecht, K.M, Buzovetsky, O, Kaderali, L, Schliemann, C, Bohnenberger, H, Angenendt, L, Hartmann, W, Wardelmann, E, Rothenburger, T, Mohr, S, Scheich, S, Comoglio, F, Wilke, A, Strobel, P, Serve, H, Michaelis, M, Ferreiros, N, Geisslinger, G, Xiong, Y, Keppler, O.T, Cinatl, J. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Selective inactivation of hypomethylating agents by SAMHD1 provides a rationale for therapeutic stratification in AML.
Nat Commun, 10, 2019
|
|
6QKI
 
 | | Native structure of EgtB from Chloracidobacterium thermophilum, a type II sulfoxide synthase | | Descriptor: | FE (III) ION, Uncharacterized protein | | Authors: | Stampfli, A.R, Badri, B.N, Schirmer, T, Seebeck, F.P. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | An Alternative Active Site Architecture for O2Activation in the Ergothioneine Biosynthetic EgtB from Chloracidobacterium thermophilum.
J.Am.Chem.Soc., 141, 2019
|
|
4H5B
 
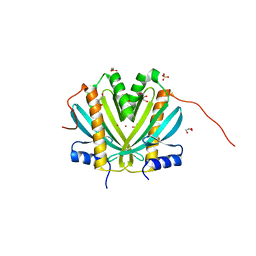 | | Crystal Structure of DR_1245 from Deinococcus radiodurans | | Descriptor: | BROMIDE ION, DR_1245 protein, GLYCEROL, ... | | Authors: | Norais, C, Servant, P, Bouthier-de-la-Tour, C, Coureux, P.D, Ithurbide, S, Vannier, F, Guerin, P, Dulberger, C.L, Satyshur, K.A, Keck, J.L, Armengaud, J, Cox, M.M, Sommer, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Deinococcus radiodurans DR1245 Protein, a DdrB Partner Homologous to YbjN Proteins and Reminiscent of Type III Secretion System Chaperones.
Plos One, 8, 2013
|
|
3NF5
 
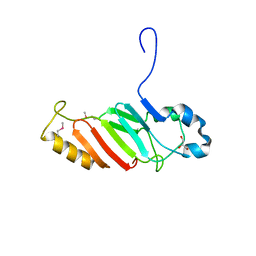 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
6Q3F
 
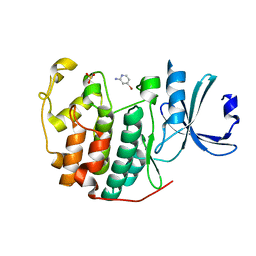 | | CDK2 in complex with FragLite2 | | Descriptor: | 4-bromanylpyridin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
4NPP
 
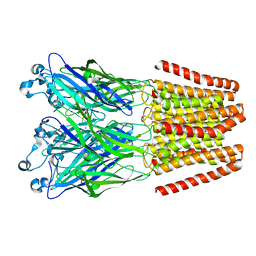 | | The GLIC-His10 wild-type structure in equilibrium between the open and locally-closed (LC) forms | | Descriptor: | NICKEL (II) ION, Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7BHE
 
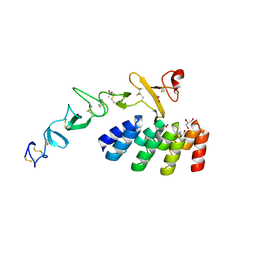 | | DARPin_D5/Her3 domain 4 complex, monoclinic crystals | | Descriptor: | ACETATE ION, DARPin_D5, GLYCEROL, ... | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7BHF
 
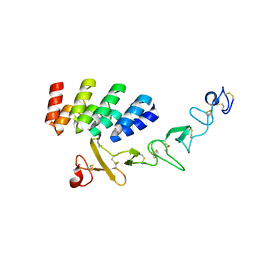 | | DARPin_D5/Her3 domain 4 complex, orthorhombic crystals | | Descriptor: | ACETATE ION, DARPin_D5, Isoform 4 of Receptor tyrosine-protein kinase erbB-3 | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6H0I
 
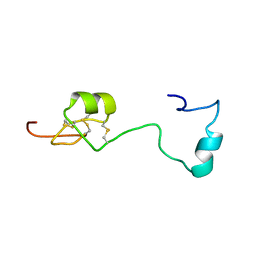 | | Solution structure of Melampsora larici-populina MlpP4.1 | | Descriptor: | Secreted protein | | Authors: | Tsan, P, Petre, B, Hecker, A, Rouhier, N, Duplessis, S. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds.
Sci Rep, 9, 2019
|
|
6RKI
 
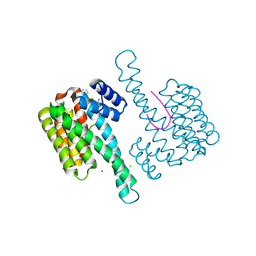 | | Fragment AZ-023 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-[(3-aminophenyl)amino]-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RKM
 
 | | Fragment AZ-022 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-phenyl-5-phenylazanyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
4ZZ6
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine triphosphate at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
