6GYZ
 
 | |
9EM8
 
 | | Oligomeric structure of SynDLP in presence of GDP | | Descriptor: | Slr0869 protein | | Authors: | Junglas, B, Gewehr, L, Schoennenbeck, P, Schneider, D, Sachse, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP.
Cell Rep, 43, 2024
|
|
8ZHD
 
 | | SARS-CoV-2 spike trimer (6P) in complex with two R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
1JO0
 
 | | Structure of HI1333, a Hypothetical Protein from Haemophilus influenzae with Structural Similarity to RNA-binding Proteins | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN HI1333 | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae
Proteins, 49, 2002
|
|
4OW6
 
 | | Crystal structure of Diphtheria Toxin at acidic pH | | Descriptor: | Diphtheria toxin | | Authors: | Leka, O, Vallese, F, Pirazzini, M, Berto, P, Montecucco, C, Zanotti, G. | | Deposit date: | 2014-01-31 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diphtheria toxin conformational switching at acidic pH.
Febs J., 281, 2014
|
|
6TSX
 
 | | Crystal structure of horse L ferritin (HoLf) Fe(III)-loaded for 30 minutes | | Descriptor: | CADMIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
1CVX
 
 | | CRYSTAL STRUCTURE OF POLYAMIDE DIMER (IMPYHPPYBETADP)2 BOUND TO B-DNA DECAMER CCAGATCTGG | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*TP*CP*TP*GP*G)-3', HYDROXYPYRROLE-IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Bremer, R.E, White, S, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural effects of DNA sequence on T.A recognition by hydroxypyrrole/pyrrole pairs in the minor groove.
J.Mol.Biol., 295, 2000
|
|
9BKI
 
 | |
1JRQ
 
 | | X-ray Structure Analysis of the Role of the Conserved Tyrosine-369 in Active Site of E. coli Amine Oxidase | | Descriptor: | CALCIUM ION, COPPER (II) ION, Copper amine oxidase | | Authors: | Murray, J.M, Kurtis, C.R, Tambarajah, W, Saysell, C.G, Wilmot, C.M, Parsons, M.R, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2001-08-14 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conserved tyrosine-369 in the active site of Escherichia coli copper amine oxidase is not essential.
Biochemistry, 40, 2001
|
|
1CF5
 
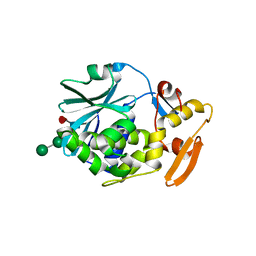 | | BETA-MOMORCHARIN STRUCTURE AT 2.55 A | | Descriptor: | PROTEIN (BETA-MOMORCHARIN), beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yuan, Y.-R, He, Y.-N, Xiong, J.-P, Xia, Z.-X. | | Deposit date: | 1999-03-24 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structure of beta-momorcharin at 2.55 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
9CJ7
 
 | | Lineage IV Lassa virus glycoprotein (Josiah) in complex with monoclonal antibody 8.9F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brouwer, P.J.M, Perrett, H.R, Ward, A.B. | | Deposit date: | 2024-07-05 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Defining bottlenecks and opportunities for Lassa virus neutralization by structural profiling of vaccine-induced polyclonal antibody responses.
Cell Rep, 43, 2024
|
|
3IPF
 
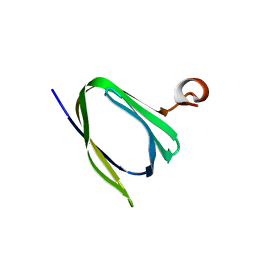 | | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR8c. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Andre, I, Rossi, P, Kennedy, M, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense.
To be Published
|
|
9CJ8
 
 | | Lineage IV Lassa virus glycoprotein (Josiah) in complex with rabbit polyclonal antibody (LAVA01-like epitope) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brouwer, P.J.M, Perrett, H.R, Ward, A.B. | | Deposit date: | 2024-07-05 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Defining bottlenecks and opportunities for Lassa virus neutralization by structural profiling of vaccine-induced polyclonal antibody responses.
Cell Rep, 43, 2024
|
|
9BK9
 
 | |
1JMJ
 
 | | Crystal Structure of Native Heparin Cofactor II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEPARIN COFACTOR II, ... | | Authors: | Baglin, T.P, Carrell, R.W, Church, F.C, Huntington, J.A. | | Deposit date: | 2001-07-18 | | Release date: | 2002-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JND
 
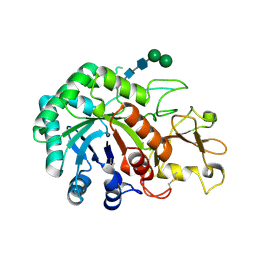 | | Crystal structure of imaginal disc growth factor-2 | | Descriptor: | Imaginal disc growth factor-2, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Varela, P.F, Llera, A.S, Mariuzza, R.A, Tormo, J. | | Deposit date: | 2001-07-23 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of imaginal disc growth factor-2. A member of a new family of growth-promoting glycoproteins from Drosophila melanogaster.
J.Biol.Chem., 277, 2002
|
|
4M4K
 
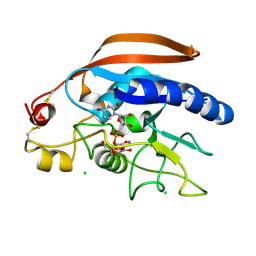 | | Crystal structure of the Drosphila beta,14galactosyltransferase 7 mutant D211N complex with manganese, UDP-Gal and xylobiose | | Descriptor: | Beta-4-galactosyltransferase 7, CHLORIDE ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of beta-1,4-Galactosyltransferase 7 Enzyme Reveal Conformational Changes and Substrate Binding.
J.Biol.Chem., 288, 2013
|
|
6YE0
 
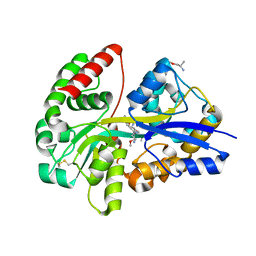 | | E.coli's Putrescine receptor PotF complexed with Putrescine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6GNM
 
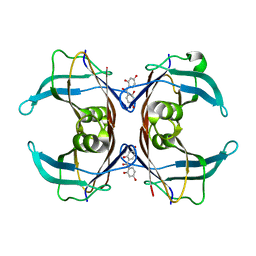 | | Crystal Structure Of Sea Bream Transthyretin in complex with 2,2',4,4'-tetrahydroxybenzophenone (BP2) | | Descriptor: | Transthyretin, bis(2,4-dihydroxyphenyl)methanone | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-05-31 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
6KM9
 
 | | Crystal structure of SucA from Vibrio vulnificus | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Seo, P.W, Kim, J.S. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Catalysis Science And Technology, 2020
|
|
9BKC
 
 | |
6GNR
 
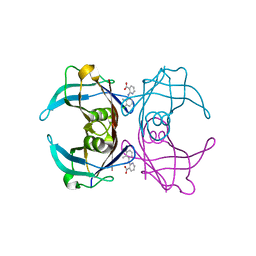 | | Crystal Structure Of Sea Bream Transthyretin in complex with 2-(3-chloro-2-methylanilino)pyridine-3-carboxylic acid (Clonixin) | | Descriptor: | 2-(3-chloro-2-methylanilino)pyridine-3-carboxylic acid, Transthyretin | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-05-31 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
6KML
 
 | | 2.09 Angstrom resolution crystal structure of tetrameric HigBA toxin-antitoxin complex from E.coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
6KMQ
 
 | | 2.3 Angstrom resolution structure of dimeric HigBA toxin-antitoxin complex from E. coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
1JP8
 
 | |
