8EU8
 
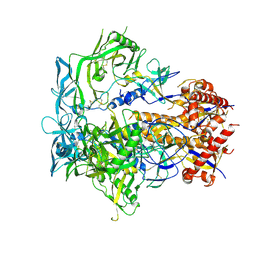 | | Cryo-EM structure of CH848 10.17DT DS-SOSIP-2P Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 10.17DT SOSIP Envelope glycoprotein gp160 | | Authors: | Wrapp, D, Acharya, P, Haynes, B.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure-Based Stabilization of SOSIP Env Enhances Recombinant Ectodomain Durability and Yield.
J.Virol., 97, 2023
|
|
3N9O
 
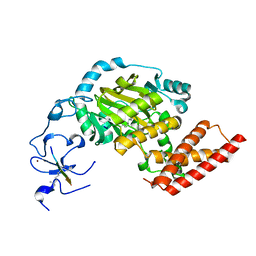 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide, H3K9me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
2V1E
 
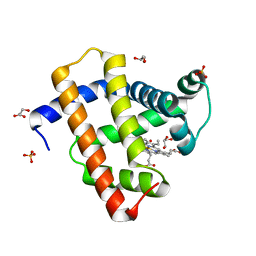 | | Crystal structure of radiation-induced myoglobin compound II - intermediate H at pH 6.8 | | Descriptor: | GLYCEROL, HYDROXIDE ION, MYOGLOBIN, ... | | Authors: | Hersleth, H.-P, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2007-05-23 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic and Spectroscopic Studies of Peroxide-Derived Myoglobin Compound II and Occurrence of Protonated Fe(Iv)-O
J.Biol.Chem., 282, 2007
|
|
2VFZ
 
 | | CRYSTAL STRUCTURE OF ALPHA-1,3 GALACTOSYLTRANSFERASE (R365K) IN COMPLEX WITH UDP-2F-GALACTOSE | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYL TRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUOROGALACTOSE | | Authors: | Jamaluddin, H, Tumbale, P, Withers, S.G, Acharya, K.R, Brew, K. | | Deposit date: | 2007-11-06 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Binding Udp-2F-Galactose to Alpha-1,3 Galactosyltransferase-Implications for Catalysis
J.Mol.Biol., 369, 2007
|
|
5KOS
 
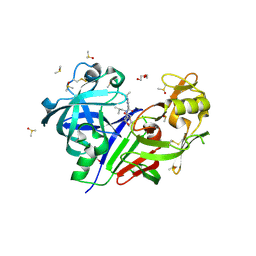 | | Discovery of TAK-272: A Novel, Potent and Orally Active Renin In-hibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-~{tert}-butyl-4-(3-methoxypropylamino)-~{N}-(2-methylpropyl)-~{N}-[(3~{S},5~{R})-5-morpholin-4-ylcarbonylpiperidin-3-yl]pyrimidine-5-carboxamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.-C, Lane, W. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of TAK-272: A Novel, Potent, and Orally Active Renin Inhibitor.
Acs Med.Chem.Lett., 7, 2016
|
|
2VIX
 
 | | Methylated Shigella flexneri MxiC | | Descriptor: | ACETATE ION, GLYCEROL, PROTEIN MXIC | | Authors: | Deane, J.E, Roversi, P, King, C, Johnson, S, Lea, S.M. | | Deposit date: | 2007-12-05 | | Release date: | 2008-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of the Shigella Flexneri Type 3 Secretion System Protein Mxic Reveal Conformational Variability Amongst Homologues.
J.Mol.Biol., 377, 2008
|
|
5NB2
 
 | |
2VK2
 
 | | Crystal structure of a galactofuranose binding protein | | Descriptor: | ABC TRANSPORTER PERIPLASMIC-BINDING PROTEIN YTFQ, beta-D-galactofuranose | | Authors: | Muller, A, Horler, R.S.P, Thomas, G.H, Wilson, K.S. | | Deposit date: | 2007-12-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Furanose-Specific Sugar Transport: Characterization of a Bacterial Galactofuranose-Binding Protein.
J.Biol.Chem., 284, 2009
|
|
8UTY
 
 | | KIF1A[1-393] P364L mutant AMP-PNP bound two-heads-bound state in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
5WX1
 
 | |
6SZ7
 
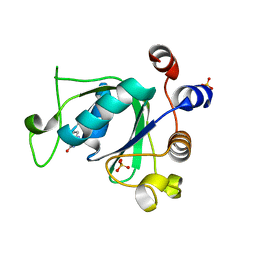 | | Crystal structure of YTHDC1 with fragment 5 (DHU_DC1_066) | | Descriptor: | 5,6,7,8-tetrahydro-4~{a}~{H}-quinazoline-2,4-dione, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
7VRP
 
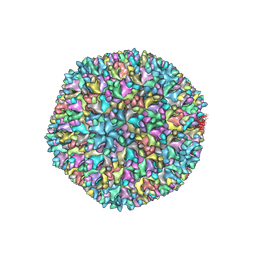 | | Structure of infectious bursal disease virus Gx strain | | Descriptor: | Structural polyprotein | | Authors: | Bao, K.Y, Zhu, P. | | Deposit date: | 2021-10-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of infectious bursal disease viruses with different virulences provide insights into their assembly and invasion
Sci Bull (Beijing), 67, 2022
|
|
6SZR
 
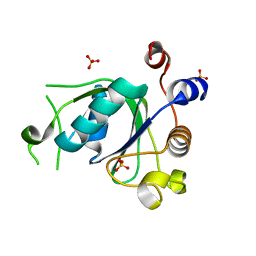 | | Crystal structure of YTHDC1 with fragment 9 (DHU_DC1_107) | | Descriptor: | 6-[[methyl-(phenylmethyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SZX
 
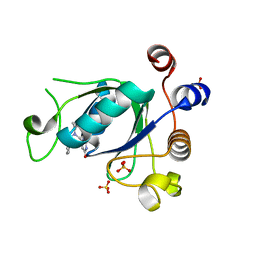 | | Crystal structure of YTHDC1 with fragment 11 (DHU_DC1_128) | | Descriptor: | 6-[[cyclopropyl-(phenylmethyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
5N5Z
 
 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation II) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
8EQ8
 
 | |
6T06
 
 | | Crystal structure of YTHDC1 with fragment 19 (DHU_DC1_045) | | Descriptor: | 3-imidazolidin-2-yl-2~{H}-indazole, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0C
 
 | | Crystal structure of YTHDC1 with fragment 26 (DHU_DC1_198) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-2~{H}-indazole-3-carboxamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
2V38
 
 | | Family 5 endoglucanase Cel5A from Bacillus agaradhaerens in complex with cellobio-derived noeuromycin | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, ENDOGLUCANASE 5A, GLYCEROL, ... | | Authors: | Gloster, T.M, Meloncelli, P.J, Money, V.A, Tarling, C.A, Davies, G.J, Withers, S.G, Stick, R.V. | | Deposit date: | 2007-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | D-Glucosylated Derivatives of Isofagomine and Noeuromycin and Their Potential as Inhibitors of Beta-Glycoside Hydrolases
To be Published
|
|
6SZM
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2009 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-methyl-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl]piperazine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
6T08
 
 | | Crystal structure of YTHDC1 with fragment 21 (DHU_DC1_131) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-(1~{H}-imidazol-2-yl)thiophene-2-sulfonamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3MXF
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective inhibition of BET bromodomains.
Nature, 468, 2010
|
|
2V53
 
 | | Crystal structure of a SPARC-collagen complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN, ... | | Authors: | Hohenester, E, Sasaki, T, Giudici, C, Farndale, R.W, Bachinger, H.P. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Sequence-Specific Collagen Recognition by Sparc.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2V8F
 
 | | Mouse Profilin IIa in complex with a double repeat from the FH1 domain of mDia1 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Kursula, I, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
2V1I
 
 | | Crystal structure of radiation-induced metmyoglobin - aqua ferrous myoglobin at pH 6.8 | | Descriptor: | GLYCEROL, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hersleth, H.-P, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2007-05-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic and Spectroscopic Studies of Peroxide-Derived Myoglobin Compound II and Occurrence of Protonated Fe(Iv)-O
J.Biol.Chem., 282, 2007
|
|
