6BK9
 
 | | Crystal Structure of Squid Arrestin | | Descriptor: | CHLORIDE ION, Visual arrestin | | Authors: | Eger, B.T, Bandyopadhyay, A, Yedidi, R.S, Ernst, O.P. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.00005579 Å) | | Cite: | A Novel Polar Core and Weakly Fixed C-Tail in Squid Arrestin Provide New Insight into Interaction with Rhodopsin.
J. Mol. Biol., 430, 2018
|
|
1GK4
 
 | | HUMAN VIMENTIN COIL 2B FRAGMENT (CYS2) | | Descriptor: | ACETATE ION, VIMENTIN | | Authors: | Strelkov, S.V, Herrmann, H, Geisler, N, Zimbelmann, R, Aebi, U, Burkhard, P. | | Deposit date: | 2001-08-08 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved Segments 1A and 2B of the Intermediate Filament Dimer: Their Atomic Structures and Role in Filament Assembly.
Embo J., 21, 2002
|
|
1I7V
 
 | | THE SOLUTION STRUCTURE OF A BAY REGION 1R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*GP*GP*AP*CP*AP*(BZA)AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Li, Z, Tamura, P.J, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2001-03-10 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (1R,2S,3R,4S)-N6-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxybenz[a]anthracenyl)]-2'-deoxyadenosyl adduct in the N-ras codon 61 sequence: DNA sequence effects
Biochemistry, 40, 2001
|
|
6WT8
 
 | | Structure of a STING-associated CdnE c-di-GMP synthase from Flavobacteriaceae sp. | | Descriptor: | STING-associated CdnE c-di-GMP synthase | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6ZEU
 
 | | Crystal structure of proteinase K lamella by electron diffraction with a 50 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.004 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
7TIM
 
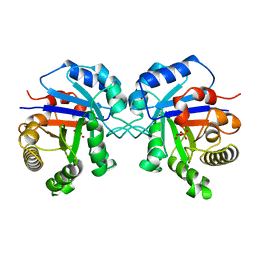 | | STRUCTURE OF THE TRIOSEPHOSPHATE ISOMERASE-PHOSPHOGLYCOLOHYDROXAMATE COMPLEX: AN ANALOGUE OF THE INTERMEDIATE ON THE REACTION PATHWAY | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Davenport, R.C, Bash, P.A, Seaton, B.A, Karplus, M, Petsko, G.A, Ringe, D. | | Deposit date: | 1991-04-23 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the triosephosphate isomerase-phosphoglycolohydroxamate complex: an analogue of the intermediate on the reaction pathway.
Biochemistry, 30, 1991
|
|
7B3I
 
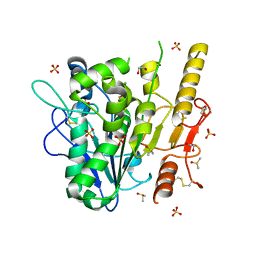 | | Notum complex with ARUK3003776 | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-chlorophenyl)sulfanyl-1$l^{4},2,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
7LQH
 
 | | Cryo-EM of KFE8 thinner nanotube (class 2, 2-sub-1) | | Descriptor: | KFE8 peptide | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Deterministic chaos in the self-assembly of beta sheet nanotubes from an amphipathic oligopeptide.
Matter, 4, 2021
|
|
7LQF
 
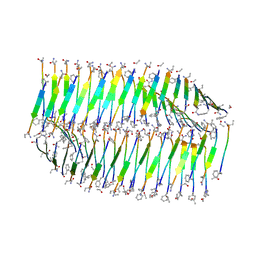 | |
6C1W
 
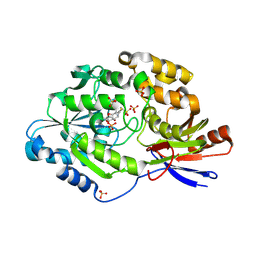 | | A tethered niacin-derived pincer complex with a nickel-carbon or sulfite-carbon bond in lactate racemase | | Descriptor: | (4S)-5-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-4-sulfo-1,4-dihydropyridine-3-carbothioic S-acid, 3-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium, Lactate racemase, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2018-01-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Lactate Racemase Nickel-Pincer Cofactor Operates by a Proton-Coupled Hydride Transfer Mechanism.
Biochemistry, 57, 2018
|
|
6ZXT
 
 | | High resolution crystal structure of chloroplastic ribose-5-phosphate isomerase from Chlamydomonas reinhardtii | | Descriptor: | Ribose-5-phosphate isomerase, SODIUM ION, SULFATE ION | | Authors: | Le Moigne, T, Crozet, P, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Crystal Structure of Chloroplastic Ribose-5-Phosphate Isomerase from Chlamydomonas reinhardtii -An Enzyme Involved in the Photosynthetic Calvin-Benson Cycle.
Int J Mol Sci, 21, 2020
|
|
7V25
 
 | |
7V28
 
 | |
7LQG
 
 | | Cryo-EM of the KFE8 thinner nanotube (class 1, C2) | | Descriptor: | KFE8 peptide | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Deterministic chaos in the self-assembly of beta sheet nanotubes from an amphipathic oligopeptide.
Matter, 4, 2021
|
|
7LQE
 
 | |
3NIU
 
 | | Crystal structure of the complex of dimeric goat lactoperoxidase with diethylene glycol at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vikram, G, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of dimeric goat lactoperoxidase with diethylene glycol at 2.9 A resolution
To be Published
|
|
6ZYX
 
 | | Outer Dynein Arm-Shulin complex - Shulin region from Tetrahymena thermophila | | Descriptor: | Dynein heavy chain, outer arm protein, Dynein intermediate chain 2, ... | | Authors: | Mali, G.R, Abid Ali, F, Lau, C.K, Carter, A.P. | | Deposit date: | 2020-08-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Shulin packages axonemal outer dynein arms for ciliary targeting.
Science, 371, 2021
|
|
7A46
 
 | | small conductance mechanosensitive channel YbiO | | Descriptor: | Putative transport protein | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6CK9
 
 | |
7AAZ
 
 | | Crystal structure of MerTK in complex with a type 1.5 aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-~{N}-[(1~{S},2~{S})-2-[[4-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]methoxy]cyclopentyl]-5-(1-methylpyrazol-4-yl)pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
411D
 
 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-METHOXYETHYL RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(T39)P*AP*CP*GP*C)-3') | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
7V35
 
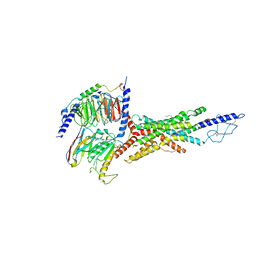 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
5GT3
 
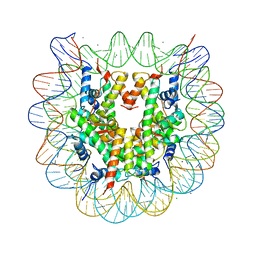 | | Crystal structure of nucleosome particle in the presence of human testis-specific histone variant, hTh2b | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-D, ... | | Authors: | Kumarevel, T, Sivaraman, P. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural analyses of the nucleosome complexes with human testis-specific histone variants, hTh2a and hTh2b
Biophys. Chem., 221, 2017
|
|
7XXF
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E},28~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26,28-dodecaen-5-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Tani, K, Kanno, R, Kurosawa, K, Takaichi, S, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-05-30 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins.
Commun Biol, 5, 2022
|
|
5Z1B
 
 | |
