4NJJ
 
 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 6-carboxy-5,6,7,8-tetrahydropterin, and Manganese(II) | | Descriptor: | (6R)-2-amino-4-oxo-3,4,5,6,7,8-hexahydropteridine-6-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
1UPX
 
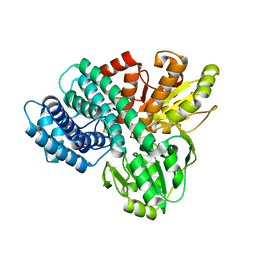 | | The crystal structure of the Hybrid Cluster Protein from Desulfovibrio desulfuricans containing molecules in the oxidized and reduced states. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE4-S3 CLUSTER, HYDROXYLAMINE REDUCTASE, ... | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Lindley, P.F. | | Deposit date: | 2003-10-14 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the Hybrid Cluster Protein (Hcp) from Desulfovibrio Desulfuricans Atcc 27774 Containing Molecules in the Oxidized and Reduced States
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FW8
 
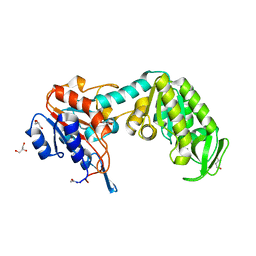 | | CIRCULARLY PERMUTED PHOSPHOGLYCERATE KINASE FROM YEAST: PGK P72 | | Descriptor: | GLYCEROL, Phosphoglycerate kinase | | Authors: | Tougard, P, Bizebard, T, Ritco-Vonsovici, M, Minard, P, Desmadril, M. | | Deposit date: | 2000-09-22 | | Release date: | 2001-03-22 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a circularly permuted phosphoglycerate kinase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4NRX
 
 | |
5I3R
 
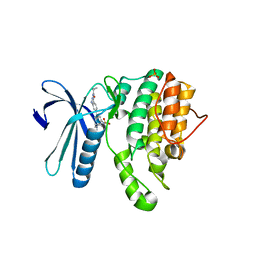 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | Descriptor: | BMP-2-inducible protein kinase, N-[6-(3-{[(cyclopropylmethyl)sulfonyl]amino}phenyl)-1H-indazol-3-yl]cyclopropanecarboxamide, PHOSPHATE ION | | Authors: | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor
To Be Published
|
|
4NXJ
 
 | | Crystal Structure of PF3D7_1475600, a bromodomain from Plasmodium Falciparum | | Descriptor: | Bromodomain protein | | Authors: | Wernimont, A.K, Loppnau, P, Knapp, S, Fonseca, M, Brennan, P.E, Dong, A, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of PF3D7_1475600, a bromodomain from Plasmodium Falciparum
TO BE PUBLISHED
|
|
3FQB
 
 | |
1V4I
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima F132A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
1URT
 
 | | MURINE CARBONIC ANHYDRASE V | | Descriptor: | CARBONIC ANHYDRASE V, ZINC ION | | Authors: | Boriack-Sjodin, P.A, Christianson, D.W. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of an intramolecular proton transfer site in murine carbonic anhydrase V.
Biochemistry, 35, 1996
|
|
4NDB
 
 | | X-ray structure of a mutant (T61D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NED
 
 | | Crystal STRUCTURE OF C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH FENOPROFEN AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Gautam, L, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-29 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal STRUCTURE OF C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH FENOPROFEN AT 2.1 ANGSTROM RESOLUTION
To be Published
|
|
1G8M
 
 | | CRYSTAL STRUCTURE OF AVIAN ATIC, A BIFUNCTIONAL TRANSFORMYLASE AND CYCLOHYDROLASE ENZYME IN PURINE BIOSYNTHESIS AT 1.75 ANG. RESOLUTION | | Descriptor: | AICAR TRANSFORMYLASE-IMP CYCLOHYDROLASE, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION | | Authors: | Greasley, S.E, Horton, P, Beardsley, G.P, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 2000-11-17 | | Release date: | 2001-04-27 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bifunctional transformylase and cyclohydrolase enzyme in purine biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
6U5H
 
 | | CryoEM Structure of Pyocin R2 - precontracted - hub | | Descriptor: | Probable bacteriophage protein Pyocin R2 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
4NJG
 
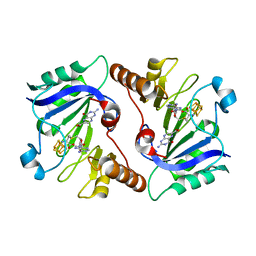 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet and 6-carboxypterin | | Descriptor: | 6-CARBOXYPTERIN, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
2Q5G
 
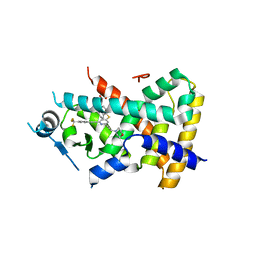 | | Ligand binding domain of PPAR delta receptor in complex with a partial agonist | | Descriptor: | Peroxisome proliferator-activated receptor delta, [(7-{[2-(3-MORPHOLIN-4-YLPROP-1-YN-1-YL)-6-{[4-(TRIFLUOROMETHYL)PHENYL]ETHYNYL}PYRIDIN-4-YL]THIO}-2,3-DIHYDRO-1H-INDEN- 4-YL)OXY]ACETIC ACID | | Authors: | Pettersson, I, Sauerberg, P, Johansson, E, Hoffman, I, Tari, L.W, Hunter, M.J, Nix, J. | | Deposit date: | 2007-06-01 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of a partial PPARdelta agonist.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2Q7C
 
 | | Crystal structure of IQN17 | | Descriptor: | CHLORIDE ION, fusion protein between yeast variant GCN4 and HIVgp41 | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Kim, P.S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
1UTC
 
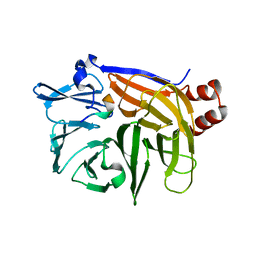 | | Clathrin terminal domain complexed with TLPWDLWTT | | Descriptor: | AMPHIPHYSIN, CLATHRIN HEAVY CHAIN | | Authors: | Miele, A.E, Evans, P.R, Owen, D.J. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller.
Nat. Struct. Mol. Biol., 11, 2004
|
|
4NR8
 
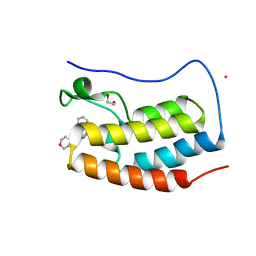 | | Crystal structure of the first bromodomain of human BRD4 in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.635 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
1UW6
 
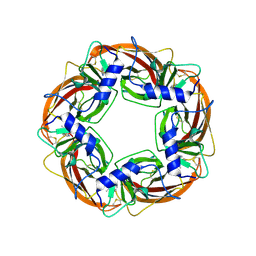 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
1V2I
 
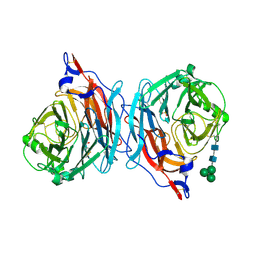 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-16 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
5A0Q
 
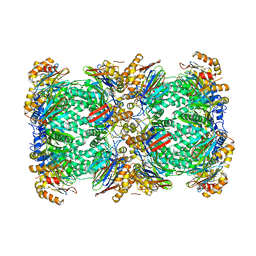 | |
5A5Q
 
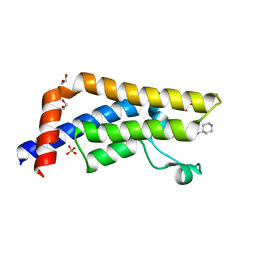 | | Crystal structure of human ATAD2 bromodomain in complex with 3-methyl- 8-piperidin-4-ylamino-1,2-dihydro-1,7-naphthyridin-2-one hydrochloride | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-8-[(piperidin-4-yl)amino]-1,2-dihydro-1,7-naphthyridin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
3T4Y
 
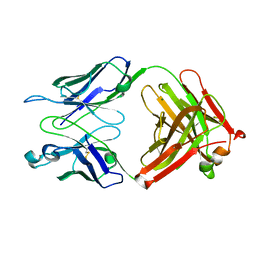 | | S25-2- KDO monosaccharide complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, MAGNESIUM ION, S25-2 FAB (IGG1K) HEAVY CHAIN, ... | | Authors: | Nguyen, H.P, Seto, N.O, Mackenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
8AJL
 
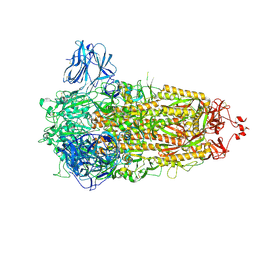 | | Structure of the Ancestral Scaffold Antigen-6 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
8AJA
 
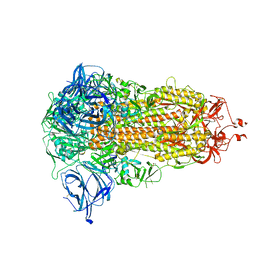 | | Structure of the Ancestral Scaffold Antigen-5 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
