7PEW
 
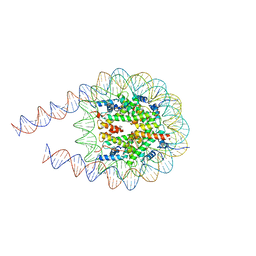 | |
7PEX
 
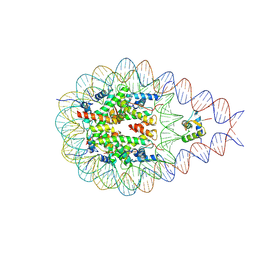 | |
7PF5
 
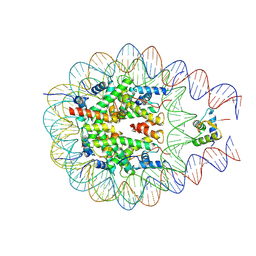 | |
5NJ4
 
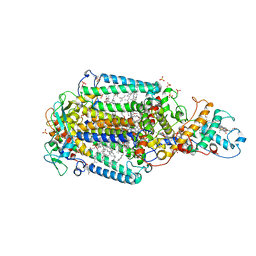 | | From macrocrystals to microcrystals: a strategy for membrane protein serial crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Dods, R, Baath, P, Branden, G, Neutze, R. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | From Macrocrystals to Microcrystals: A Strategy for Membrane Protein Serial Crystallography.
Structure, 25, 2017
|
|
7PF6
 
 | |
6VVW
 
 | |
7PFD
 
 | |
7PFE
 
 | |
7PFV
 
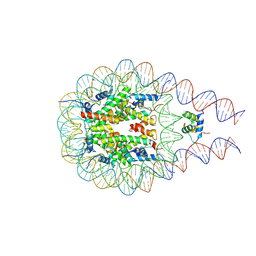 | |
7PFW
 
 | |
6ZI9
 
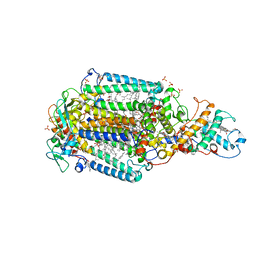 | | Ultrafast Structural Response to Charge Redistribution Within a Photosynthetic Reaction Centre - 300 ps (b) structure | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baath, P, Dods, R, Braenden, G, Neutze, R. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ultrafast structural changes within a photosynthetic reaction centre.
Nature, 589, 2021
|
|
7AJQ
 
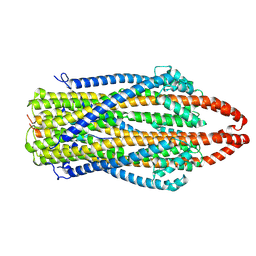 | | cryo-EM structure of ExbBD from Serratia Marcescens | | Descriptor: | Biopolymer transport protein ExbB, Biopolymer transport protein ExbD | | Authors: | Biou, V, Adaixo, R, Coureux, P.D, Delepelaire, P, Chami, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Commun Biol, 5, 2022
|
|
7Z6O
 
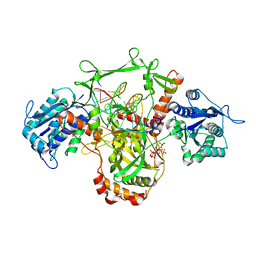 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Varela, P.F, Charbonnier, J.B. | | Deposit date: | 2022-03-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
8P4C
 
 | | Structural insights into human co-transcriptional capping - structure 3 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4D
 
 | | Structural insights into human co-transcriptional capping - structure 4 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
6W8H
 
 | | H-PGDS complexed with inhibitor 1Y | | Descriptor: | 1,2-ETHANEDIOL, 7-cyclopropyl-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,8-naphthyridine-3-carboxamide, GLUTATHIONE, ... | | Authors: | Shewchuk, L.M, Ward, P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The exploration of aza-quinolines as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors with low brain exposure.
Bioorg.Med.Chem., 28, 2020
|
|
7NAU
 
 | | Bacterial 30S ribosomal subunit assembly complex state C (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAV
 
 | | Bacterial 30S ribosomal subunit assembly complex state D (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAT
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAX
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state I (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAR
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state F (+RsgA)(Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7PHO
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 7.1 in complex with 4-hydroxybenzaldehyde | | Descriptor: | BICARBONATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Nunes-Costa, D, Silva, A, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-08-17 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 7.1 in complex with 4-hydroxybenzaldehyde
To Be Published
|
|
8EL2
 
 | | SARS-CoV-2 RBD bound to neutralizing antibody Fab ICO-hu23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ICO-hu23 Heavy Chain, Fab ICO-hu23 Light Chain, ... | | Authors: | Besaw, J.E, Kuo, A, Morizumi, T, Ernst, O.P. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
7Q5W
 
 | | The tandem SH2 domains of SYK with a bound TYROBP diphospho-ITAM peptide | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Chen, Z, Bountra, C, von Delft, F, Gileadi, O, Brennan, P.E. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The tandem SH2 domains of SYK
To Be Published
|
|
8SCB
 
 | | Terminating ribosome with SRI-41315 | | Descriptor: | (2S,4aS)-2-cyclobutyl-10-methyl-3-phenyl-2,10-dihydropyrimido[4,5-b]quinoline-4,5(3H,4aH)-dione, 18S_rRNA, 28S_rRNA, ... | | Authors: | Yip, M.C.J, Coelho, J.P.L, Oltion, K, Tauton, J, Shao, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center.
Nat.Chem.Biol., 20, 2024
|
|
