3I7S
 
 | | Dihydrodipicolinate synthase mutant - K161A - with the substrate pyruvate bound in the active site. | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
4HIN
 
 | | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L) | | Descriptor: | COPPER (II) ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L)
To be Published
|
|
1F0S
 
 | | Crystal Structure of Human Coagulation Factor XA Complexed with RPR208707 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XA, THIENO[3,2-B]PYRIDINE-2-SULFONIC ACID [2-OXO-1-(1H-PYRROLO[2,3-C]PYRIDIN-2-YLMETHYL)-PYRROLIDIN-3-YL]-AMIDE | | Authors: | Maignan, S, Guilloteau, J.P, Pouzieux, S, Choi-Sledeski, Y.M, Becker, M.R, Klein, S.I, Ewing, W.R, Pauls, H.W, Spada, A.P, Mikol, V. | | Deposit date: | 2000-05-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human factor Xa complexed with potent inhibitors.
J.Med.Chem., 43, 2000
|
|
1P1V
 
 | | Crystal Structure of FALS-associated human Copper-Zinc Superoxide Dismutase (CuZnSOD) Mutant D125H to 1.4A | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Malek, K, Rodriguez, J.A, Doucette, P.A, Taylor, A.B, Hayward, L.J, Cabelli, D.E, Valentine, J.S, Hart, P.J. | | Deposit date: | 2003-04-14 | | Release date: | 2003-08-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Alternative Mechanism of Bicarbonate-mediated Peroxidation by Copper-Zinc Superoxide Dismutase: RATES ENHANCED VIA PROPOSED ENZYME-ASSOCIATED PEROXYCARBONATE INTERMEDIATE
J.Biol.Chem., 278, 2003
|
|
2IZM
 
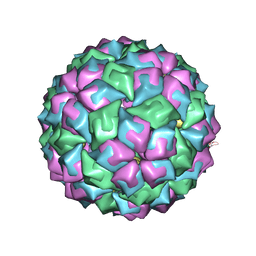 | | MS2-RNA HAIRPIN (C-10) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*CP*GP*GP*AP*UP *CP*AP*CP*CP*CP*AP*UP*GP*U)-3', Capsid protein | | Authors: | Helgstrand, C, Grahn, E, Moss, T, Stonehouse, N.J, Tars, K, Stockley, P.G, Liljas, L. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Investigating the Structural Basis of Purine Specificity in the Structures of MS2 Coat Protein RNA Translational Operator Complexes
Nucleic Acids Res., 30, 2002
|
|
1G09
 
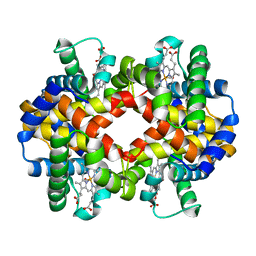 | | CARBONMONOXY LIGANDED BOVINE HEMOGLOBIN PH 7.2 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
2IGG
 
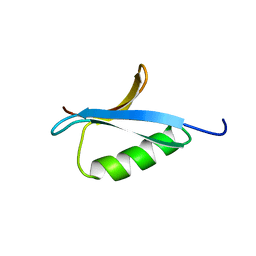 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
1SF2
 
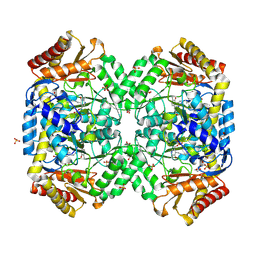 | | Structure of E. coli gamma-aminobutyrate aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
4I05
 
 | | Structure of intermediate processing form of cathepsin B1 from Schistosoma mansoni | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2012-11-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activation route of the Schistosoma mansoni cathepsin B1 drug target: structural map with a glycosaminoglycan switch
Structure, 22, 2014
|
|
1GAJ
 
 | | CRYSTAL STRUCTURE OF A NUCLEOTIDE-FREE ATP-BINDING CASSETTE FROM AN ABC TRANSPORTER | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HIGH-AFFINITY BRANCHED CHAIN AMINO ACID TRANSPORT ATP-BINDING PROTEIN, ... | | Authors: | Karpowich, N, Yuan, Y.-R, Dai, P.L, Martsinkevich, O, Millen, L, Thomas, P.J, Hunt, J.F. | | Deposit date: | 2000-11-30 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter.
Structure, 9, 2001
|
|
5U3Q
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 1 | | Descriptor: | 6-(2-{[([1,1'-biphenyl]-4-carbonyl)(propan-2-yl)amino]methyl}phenoxy)hexanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3IRN
 
 | | Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase COMPLEXED WITH NADPH AND Cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2009-08-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of antifolate inhibition of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase
To be Published
|
|
2J7V
 
 | | Structure of PBP-A | | Descriptor: | TLL2115 PROTEIN | | Authors: | Evrard, C, Declercq, J.P. | | Deposit date: | 2006-10-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pbp-A from Thermosynechococcus Elongatus, a Penicillin-Binding Protein Closely Related to Class a Beta-Lactamases.
J.Mol.Biol., 386, 2009
|
|
1ONZ
 
 | | Oxalyl-aryl-Amino Benzoic acid Inhibitors of PTP1B, compound 8b | | Descriptor: | 2-[(7-HYDROXY-NAPHTHALEN-1-YL)-OXALYL-AMINO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
7JJM
 
 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS | | Descriptor: | CHLORIDE ION, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McGuaig, R, Tan, H.Y.A, Hardy, C, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
4HKT
 
 | |
1P4T
 
 | | Crystal structure of Neisserial surface protein A (NspA) | | Descriptor: | ETHANOLAMINE, PENTAETHYLENE GLYCOL MONODECYL ETHER, SULFATE ION, ... | | Authors: | Vandeputte-Rutten, L, Bos, M.P, Tommassen, J, Gros, P. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Neisserial Surface Protein A (NspA),
a conserved outer membrane protein with vaccine potential
J.Biol.Chem., 278, 2003
|
|
1SJ0
 
 | | Human Estrogen Receptor Alpha Ligand-binding Domain in Complex with the Antagonist Ligand 4-D | | Descriptor: | (2S,3R)-2-(4-(2-(PIPERIDIN-1-YL)ETHOXY)PHENYL)-2,3-DIHYDRO-3-(4-HYDROXYPHENYL)BENZO[B][1,4]OXATHIIN-6-OL, Estrogen receptor | | Authors: | Kim, S, Wu, J.Y, Birzin, E.T, Chan, W, Pai, L.Y, Yang, Y.T, Mosley, R.T, Fitzgerald, P.M, Sharma, N, DiNinno, F, Rohrer, S.P, Schaeffer, J.M, Hammond, M.L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estrogen Receptor Ligands. II. Discovery of Benzoxathiins as Potent, Selective Estrogen Receptor alpha Modulators.
J.Med.Chem., 47, 2004
|
|
1P5N
 
 | |
5UDT
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Lactate racemization operon protein LarE, PHOSPHATE ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4M8D
 
 | | Crystal structure of an isatin hydrolase bound to product analogue thioisatinate | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Putative uncharacterized protein, ... | | Authors: | Bjerregaard-Andersen, K, Sommer, T, Jensen, J.K, Jochimsen, B, Etzerodt, M, Morth, J.P. | | Deposit date: | 2013-08-13 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A proton wire and water channel revealed in the crystal structure of isatin hydrolase.
J.Biol.Chem., 289, 2014
|
|
3J07
 
 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2MIX
 
 | | Structure of a novel venom peptide toxin from sample limited terebrid marine snail | | Descriptor: | venom peptide toxin | | Authors: | Bhuiyan, M.H, Anand, P, Grigoryan, A, Holford, M, Poget, S.F. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sample limited characterization of a novel disulfide-rich venom peptide toxin from terebrid marine snail Terebra variegata.
Plos One, 9, 2014
|
|
2HBB
 
 | | Crystal Structure of the N-terminal Domain of Ribosomal Protein L9 (NTL9) | | Descriptor: | 50S ribosomal protein L9, ZINC ION | | Authors: | Cho, J.-H, Kim, E.Y, Schindelin, H, Raleigh, D.P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energetically significant networks of coupled interactions within an unfolded protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7JFQ
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
