6L9D
 
 | | X-ray structure of synthetic GB1 domain with mutations K10(DVA), T11S | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Penmatsa, A, Chatterjee, J, Majumder, P, Khatri, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Increasing protein stability by engineering the n -> pi * interaction at the beta-turn.
Chem Sci, 11, 2020
|
|
4U02
 
 | | Crystal structure of apo-TTHA1159 | | Descriptor: | Amino acid ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | Karthiga Devi, S, Chichili, V.P.R, Velmurugan, D, Sivaraman, J. | | Deposit date: | 2014-07-11 | | Release date: | 2015-05-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for the hydrolysis of ATP by a nucleotide binding subunit of an amino acid ABC transporter from Thermus thermophilus
J.Struct.Biol., 190, 2015
|
|
7QH5
 
 | |
4U4L
 
 | | Crystal structure of the metallo-beta-lactamase NDM-1 in complex with a bisthiazolidine inhibitor | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase NDM-1, GLYCEROL, ... | | Authors: | Kosmopoulou, M, Hinchliffe, P, Spencer, J. | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase NDM-1 in complex with a bisthiazolidine inhibitor
To Be Published
|
|
4U1E
 
 | | Crystal structure of the eIF3b-CTD/eIF3i/eIF3g-NTD translation initiation complex | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit G, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Zhang, S, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell, 158, 2014
|
|
7QSG
 
 | |
7QTE
 
 | | Crystal Structure of the Fe(II)/alpha-ketoglutarate dependent dioxygenase PlaO1 in complex with cobalt and succinate | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, COBALT (II) ION, PlaO1, ... | | Authors: | Lukat, P, Daum, M, Bechthold, A, Einsle, O. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural investigations on the Fe(II)/alpha-ketoglutarate dependent dioxygense PlaO1 from Streptomyces sp. Tu6071
Thesis, 2011
|
|
4U46
 
 | | Crystal structure of an avidin mutant | | Descriptor: | Avidin, CHLORIDE ION | | Authors: | Agrawal, N, Lehtonen, S, Kahkonen, N, Riihimaki, T, Hytonen, V.P, Kulomaa, M.S, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Artificial Avidin-Based Receptors for a Panel of Small Molecules.
Acs Chem.Biol., 11, 2016
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
7QRI
 
 | | Regulatory domain dimer of tryptophan hydroxylase 2 in complex with L-Phe | | Descriptor: | PHENYLALANINE, Tryptophan 5-hydroxylase 2 | | Authors: | Vedel, I.M, Prestel, A, Harris, P, Peters, G.H.J, Kragelund, B.B. | | Deposit date: | 2022-01-11 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of human tryptophan hydroxylase 2 reveals that L-Phe is superior to L-Trp as the regulatory domain ligand.
Structure, 31, 2023
|
|
7QUH
 
 | | Siglec-8 in complex with therapeutic Fab AK002. | | Descriptor: | Sialic acid-binding Ig-like lectin 8, Sialic acid-binding immunoglobulin-type lectin | | Authors: | Lenza, M.P, Oyenarte, I, Jimenez Barbero, J, Ereno Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
7QA5
 
 | |
7QP5
 
 | | Crystal Structure of E. coli FhuF | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferric iron reductase protein FhuF | | Authors: | Trindade, I.B, Rollo, F, Matias, P.M, Moe, E, Louro, R.O. | | Deposit date: | 2022-01-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of a novel ferredoxin: FhuF, a ferric-siderophore reductase from E. coli K-12 with a novel 2Fe-2S cluster coordination
Biorxiv, 2023
|
|
4UAM
 
 | | 1.8 Angstrom crystal structure of IMP-1 metallo-beta-lactamase with a mixed iron-zinc center in the active site | | Descriptor: | CITRATE ANION, FE (III) ION, IMP-1 metallo-beta-lactamase, ... | | Authors: | Carruthers, T.J, Carr, P.D, Jackson, C.J, Otting, G. | | Deposit date: | 2014-08-11 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Iron(III) Located in the Dinuclear Metallo-beta-Lactamase IMP-1 by Pseudocontact Shifts.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4TWT
 
 | | Human TNFa dimer in complex with the semi-synthetic bicyclic peptide M21 | | Descriptor: | (2,4,6-trimethylbenzene-1,3,5-triyl)trimethanol, ALA-CYS-PRO-PRO-CYS-LEU-TRP-GLN-VAL-LEU-CYS-GLY, GLYCEROL, ... | | Authors: | Luzi, S, Kondo, Y, Bernard, E, Stadler, L, Winter, G, Holliger, P. | | Deposit date: | 2014-07-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Subunit disassembly and inhibition of TNF alpha by a semi-synthetic bicyclic peptide.
Protein Eng.Des.Sel., 28, 2015
|
|
4U63
 
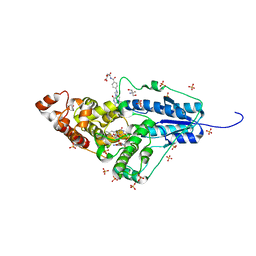 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
4U9W
 
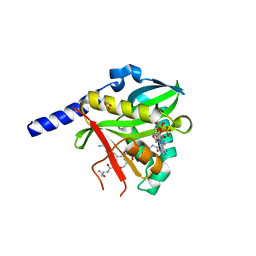 | | Crystal Structure of NatD bound to H4/H2A peptide and CoA | | Descriptor: | COENZYME A, GLYCEROL, Histone H4/H2A N-terminus, ... | | Authors: | Magin, R.S, Liszczak, G.P, Marmorstein, R. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Molecular Basis for Histone H4- and H2A-Specific Amino-Terminal Acetylation by NatD.
Structure, 23, 2015
|
|
7RFA
 
 | |
4UD8
 
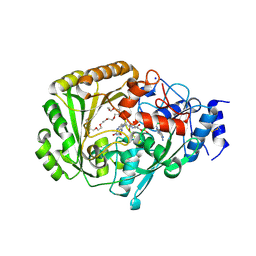 | | AtBBE15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Daniel, B, Steiner, B, Pavkov-Keller, T, Dordic, A, Gutmann, A, Sensen, C.W, Nidetzky, B, van der Graaff, E, Wallner, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Oxidation of Monolignols by Members of the Berberine Bridge Enzyme Family Suggests a Role in Cell Wall Metabolism.
J.Biol.Chem., 290, 2015
|
|
4UMN
 
 | | Structure of a stapled peptide antagonist bound to Nutlin-resistant Mdm2. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, M06 | | Authors: | Chee, S, Wongsantichon, J, Quah, S, Robinson, R.C, Verma, C, Lane, D.P, Brown, C.J, Ghadessy, F.J. | | Deposit date: | 2014-05-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
PLoS ONE, 9, 2014
|
|
4UP1
 
 | |
4UHK
 
 | |
4UP2
 
 | | Crystal structure of Escherichia coli tryptophanase purified from alkaline stressed bacterial culture. | | Descriptor: | BETA-MERCAPTOETHANOL, SULFATE ION, TRYPTOPHANASE | | Authors: | Rety, S, Deschamps, P, Leulliot, N. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of Escherichia Coli Tryptophanase Purified from an Alkaline-Stressed Bacterial Culture.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4UOO
 
 | |
4UD7
 
 | | Structure of the stapled peptide YS-02 bound to MDM2 | | Descriptor: | MDM2, YS-02 | | Authors: | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | Deposit date: | 2014-12-08 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
