5O41
 
 | | Low-dose fixed target serial synchrotron crystallography structure of sperm whale myoglobin | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Owen, R.L, Axford, D, Sherrell, D, Mueller-Werkmeister, H. | | Deposit date: | 2017-05-25 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low-dose fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
9EPE
 
 | | 8fs pulse duration 10uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-18 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQY
 
 | | 41fs pulse duration 100uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQZ
 
 | | 53fs pulse duration 10uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9ER0
 
 | | 53fs pulse duration 50uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9ER1
 
 | | 53fs pulse duration 100uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQR
 
 | | 8fs pulse duration 50uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQV
 
 | | 41fs pulse duration 10uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQS
 
 | | 8fs pulse duration 100uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQU
 
 | | 24fs pulse duration 100uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQT
 
 | | 24fs pulse duration 10uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
9EQX
 
 | | 41fs pulse duration 50uJ pulse energy thaumatin | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Owen, R.L, Hough, M.A, Worrall, J, Williams, L. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Damage before destruction? X-ray-induced changes in single-pulse serial femtosecond crystallography.
Iucrj, 12, 2025
|
|
4N6U
 
 | | Adhiron: a stable and versatile peptide display scaffold - truncated adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
7QT8
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound ABT-957 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QT6
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z1367324110 | | Descriptor: | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QT9
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z4439011584 | | Descriptor: | DIMETHYL SULFOXIDE, N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide, Non-structural protein 6 | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QT5
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z31792168 | | Descriptor: | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QT7
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z4439011520 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(5-tert-butyl-1,2-oxazol-3-yl)-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5ML9
 
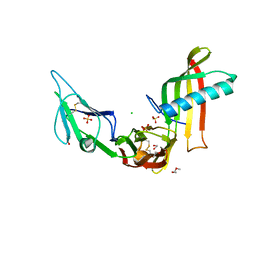 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MN2
 
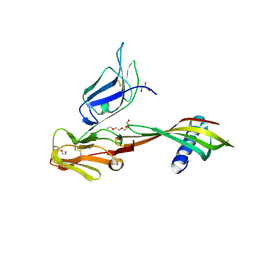 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4N6T
 
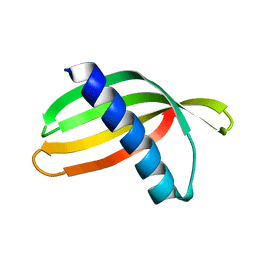 | | Adhiron: a stable and versatile peptide display scaffold - full length adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
2WUG
 
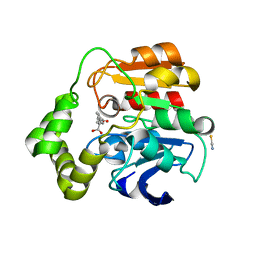 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R.L, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a C-C Hydrolase from Mycobacterium Tuberuclosis Involved in Cholesterol Metabolism.
J.Biol.Chem., 285, 2010
|
|
7ZMJ
 
 | | SFX structure of dye-type peroxidase DtpB R243A variant in the ferric state | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Putative dye-decolorizing peroxidase (DyP), ... | | Authors: | Lucic, M, Worrall, J.A.R, Hough, M.A, Shilova, A, Axford, D.A, Owen, R.L, Tosha, T, Sugimoto, H, Owada, S. | | Deposit date: | 2022-04-19 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serial Femtosecond Crystallography Reveals the Role of Water in the One- or Two-Electron Redox Chemistry of Compound I in the Catalytic Cycle of the B-Type Dye-Decolorizing Peroxidase DtpB.
Acs Catalysis, 12, 2022
|
|
4M92
 
 | | Crystal structure of hN33/Tusc3-peptide 2 | | Descriptor: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M91
 
 | | crystal structure of hN33/Tusc3-peptide 1 | | Descriptor: | Protein cereblon, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
