2MCX
 
 | | Solid-state NMR structure of piscidin 3 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Wieczorek, W.E, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2021-08-18 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MCU
 
 | | Solid-state NMR structure of piscidin 1 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
1F11
 
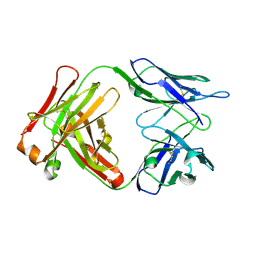 | | F124 FAB FRAGMENT FROM A MONOCLONAL ANTI-PRES2 ANTIBODY | | Descriptor: | F124 IMMUNOGLOBULIN (IGG1 HEAVY CHAIN), F124 IMMUNOGLOBULIN (KAPPA LIGHT CHAIN) | | Authors: | Saul, F.A, Vulliez-Le Normand, B, Passafiume, M, Riottot, M.M, Bentley, G.A. | | Deposit date: | 2000-05-18 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Fab fragment from F124, a monoclonal antibody specific for hepatitis B surface antigen.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2OR1
 
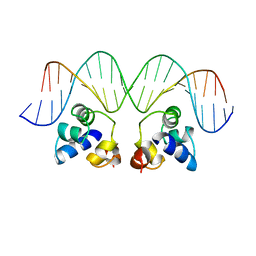 | | RECOGNITION OF A DNA OPERATOR BY THE REPRESSOR OF PHAGE 434. A VIEW AT HIGH RESOLUTION | | Descriptor: | 434 REPRESSOR, DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3') | | Authors: | Aggarwal, A.K, Rodgers, D.W, Drottar, M, Ptashne, M, Harrison, S.C. | | Deposit date: | 1989-09-05 | | Release date: | 1989-09-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of a DNA operator by the repressor of phage 434: a view at high resolution.
Science, 242, 1988
|
|
4BIH
 
 | | Crystal structure of the conserved staphylococcal antigen 1A, Csa1A | | Descriptor: | CALCIUM ION, UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
1YZW
 
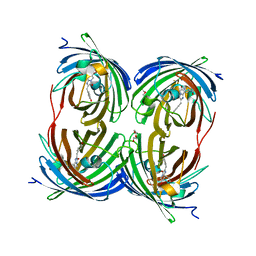 | | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore | | Descriptor: | DI(HYDROXYETHYL)ETHER, GFP-like non-fluorescent chromoprotein | | Authors: | Wilmann, P.G, Petersen, J, Pettikiriarachchi, A, Buckle, A.M, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2005-02-28 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore
J.Mol.Biol., 349, 2005
|
|
5OJ9
 
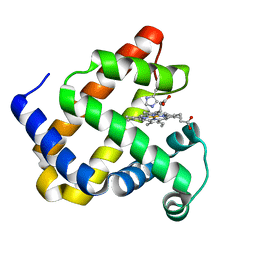 | | Structure of Mb NMH | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
1PCI
 
 | | PROCARICAIN | | Descriptor: | PROCARICAIN | | Authors: | Groves, M.R, Taylor, M.A.J, Scott, M, Cummings, N.J, Pickersgill, R.W, Jenkins, J.A. | | Deposit date: | 1996-06-28 | | Release date: | 1997-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The prosequence of procaricain forms an alpha-helical domain that prevents access to the substrate-binding cleft.
Structure, 4, 1996
|
|
2P4M
 
 | | High pH structure of Rtms5 H146S variant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Wilmann, P.G, Olsen, S, Byres, E, Smith, S.C, Dove, S.G, Turcic, K.N, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for the pH-dependent increase in fluorescence efficiency of chromoproteins
J.Mol.Biol., 368, 2007
|
|
4BIG
 
 | | Crystal structure of the conserved staphylococcal antigen 1B, Csa1B | | Descriptor: | UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4NAL
 
 | | Arabidopsis thaliana IspD in complex with tribromodichloro-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAI
 
 | | Arabidopsis thaliana IspD apo | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAK
 
 | | Arabidopsis thaliana IspD in complex with pentabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAN
 
 | | Arabidopsis thaliana IspD in complex with tetrabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5OJC
 
 | | Structure of MbQ2.1 NMH | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
2J5L
 
 | | Structure of a Plasmodium falciparum apical membrane antigen 1-Fab F8. 12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
5OJA
 
 | | Structure of MbQ | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
2JW6
 
 | | Solution structure of the DEAF1 MYND domain | | Descriptor: | Deformed epidermal autoregulatory factor 1 homolog, ZINC ION | | Authors: | Spadaccini, R, Perrin, H, Bottomley, M, Ansieu, S, Sattler, M. | | Deposit date: | 2007-10-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Retraction notice to "Structure and functional analysis of the MYND domain" [J. Mol. Biol. (2006) 358, 498-508].
J.Mol.Biol., 376, 2008
|
|
3VIC
 
 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Byres, E, Wilce, M, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
3VK1
 
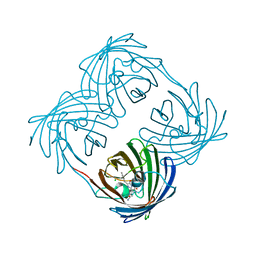 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Wilce, M, Byres, M, Rossjohn, J, Devenish, R.J, Prescott, M. | | Deposit date: | 2011-11-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
1D8B
 
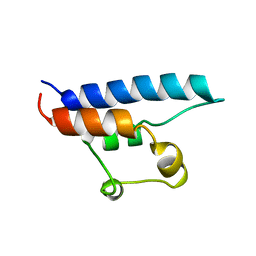 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
5OJB
 
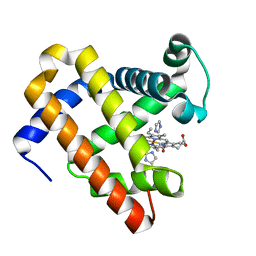 | | Structure of MbQ NMH | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
3RKZ
 
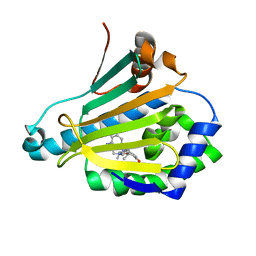 | | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor capable of significantly decreasing tumor volume in a mouse xenograft model. | | Descriptor: | (5R,6S)-3-(L-alanyl)-5,6,15,15,18-pentamethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-12,8-(metheno)[1,5,9]triazacyclotetradecino[1,2-a]indole-9-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, Li, Z, Dushin, R.G, Nittoli, T, Otteng, M, Nikitenko, A, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E, Olland, A, Johnson, M, Levin, J.I. | | Deposit date: | 2011-04-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5693 Å) | | Cite: | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor which significantly decreases tumor volume in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4DMW
 
 | | Crystal structure of the GT domain of Clostridium difficile toxin A (TcdA) in complex with UDP and Manganese | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Scarselli, M, Maione, D, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
4DMV
 
 | | Crystal structure of the GT domain of Clostridium difficile Toxin A | | Descriptor: | Toxin A | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Maione, D, Scarselli, M, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
