8JHH
 
 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | Descriptor: | GLYCEROL, MnLam55A | | Authors: | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
6M5W
 
 | | Co-crystal structure of human serine hydroxymethyltransferase 1 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
6M5O
 
 | | Co-crystal structure of human serine hydroxymethyltransferase 2 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, Serine hydroxymethyltransferase, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30000663 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
5XFW
 
 | | Crystal structures of FMN-free form of dihydroorotate dehydrogenase from Trypanosoma brucei | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), MALONATE ION | | Authors: | Kubota, T, Tani, O, Yamaguchi, T, Namatame, I, Sakashita, H, Furukawa, K, Yamasaki, K. | | Deposit date: | 2017-04-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of FMN-bound and FMN-free forms of dihydroorotate dehydrogenase fromTrypanosoma brucei.
FEBS Open Bio, 8, 2018
|
|
5XFV
 
 | | Crystal structures of FMN-bound form of dihydroorotate dehydrogenase from Trypanosoma brucei | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, MALONATE ION | | Authors: | Kubota, T, Tani, O, Yamaguchi, T, Namatame, I, Sakashita, H, Furukawa, K, Yamasaki, K. | | Deposit date: | 2017-04-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of FMN-bound and FMN-free forms of dihydroorotate dehydrogenase fromTrypanosoma brucei.
FEBS Open Bio, 8, 2018
|
|
2D7R
 
 | | Crystal structure of pp-GalNAc-T10 complexed with GalNAc-Ser on lectin domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
2D7I
 
 | | Crystal structure of pp-GalNAc-T10 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
3ASQ
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with H-antigen | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASR
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-a | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASS
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASP
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with A-antigen | | Descriptor: | Capsid protein, SODIUM ION, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3AST
 
 | | Crystal structure of P domain Q389N mutant from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
1JL8
 
 | | Complex of alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with beta-cyclodextrin based on a co-crystallization with methyl beta-cyclodextrin | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1JIB
 
 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1X3X
 
 | | Crystal Structure of Cytochrome b5 from Ascaris suum | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yokota, T, Nakajima, Y, Yamakura, F, Sugio, S, Hashimoto, M, Takamiya, S, Aoki, T. | | Deposit date: | 2005-05-11 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique structure of Ascaris suum b5-type cytochrome: an additional alpha-helix and positively charged residues on the surface domain interact with redox partners
Biochem.J., 394, 2006
|
|
1WRM
 
 | | Crystal structure of JSP-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, dual specificity phosphatase 22 | | Authors: | Yokota, T, Kashima, A, Kato, R, Sugio, S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-10-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human dual specificity phosphatase, JNK stimulatory phosphatase-1, at 1.5 A resolution
Proteins, 66, 2006
|
|
8H25
 
 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and Structure of Lacticaseibacillus casei GH35 beta-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lacto- N -biose I and Galacto- N -biose.
J Appl Glycosci (1999), 70, 2023
|
|
1QZM
 
 | | alpha-domain of ATPase | | Descriptor: | ATP-dependent protease La | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Khalatova, A.G, Rasulova, F.S, Tropea, J.E, Maurizi, M.R, Rotanova, T.V, Gustchina, A, Wlodawer, A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the AAA+ alpha domain of E. coli Lon protease at 1.9A resolution.
J.Struct.Biol., 146
|
|
3C9D
 
 | | Crystal structure of Vps75 | | Descriptor: | Vacuolar protein sorting-associated protein 75 | | Authors: | Berndsen, C.E, Tsubota, T, Lindner, S.E, Lee, S, Holton, J.M, Kaufman, P.D, Keck, J.L, Denu, J.M. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75
Nat.Struct.Mol.Biol., 15, 2008
|
|
7MS7
 
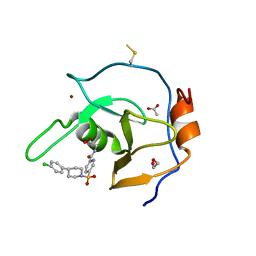 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS5
 
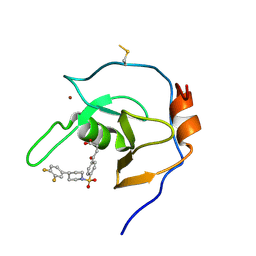 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS6
 
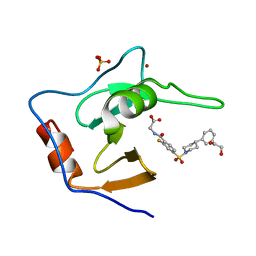 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6LK4
 
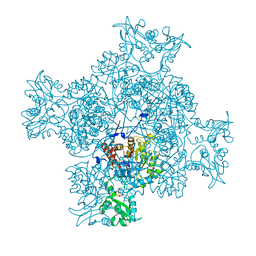 | | Crystal structure of GMP reductase from Trypanosoma brucei in complex with guanosine 5'-triphosphate | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Guanosine 5'-monophosphate Reductase, PHOSPHATE ION | | Authors: | Mase, H, Otani, T, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-12-18 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
6LFF
 
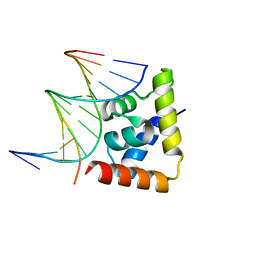 | | transcription factor SATB1 CUTr1 domain in complex with a phosphorothioate DNA | | Descriptor: | DNA (5'-D(*GP*(C7R)P*(PST)P*AP*AP*TP*AP*TP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*(AS)P*(PST)P*(AS)P*(PST)P*TP*AP*GP*C)-3'), DNA-binding protein SATB1 | | Authors: | Akutsu, Y, Kubota, T, Yamasaki, T, Yamasaki, K. | | Deposit date: | 2019-12-02 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Enhanced affinity of racemic phosphorothioate DNA with transcription factor SATB1 arising from diastereomer-specific hydrogen bonds and hydrophobic contacts.
Nucleic Acids Res., 48, 2020
|
|
6LUE
 
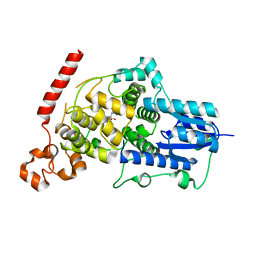 | | Crystal structure of mouse Cryptochrome 1 in complex with compound KL201 | | Descriptor: | 2-bromanyl-N-(5,6,7,8-tetrahydro-[1]benzothiolo[2,3-d]pyrimidin-4-yl)benzamide, Cryptochrome-1 | | Authors: | Miller, S, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Isoform-Selective Modulator of Cryptochrome 1 Regulates Circadian Rhythms in Mammals.
Cell Chem Biol, 27, 2020
|
|
