292D
 
 | | INTERACTION BETWEEN THE LEFT-HANDED Z-DNA AND POLYAMINE:THE CRYSTAL STRUCTURE OF THE D(CG)3 AND N-(2-AMINOETHYL)-1,4-DIAMINOBUTANE COMPLEX | | Descriptor: | 1-(AMINOETHYL)AMINO-4-AMINOBUTANE, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Ohishi, H, Kunisawa, S, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Tomita, K, Hakoshima, T. | | Deposit date: | 1991-10-09 | | Release date: | 1996-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between the left-handed Z-DNA and polyamine. The crystal structure of the d(CG)3 and N-(2-aminoethyl)-1,4-diamino-butane complex.
FEBS Lett., 284, 1991
|
|
5B5W
 
 | | Crystal structure of MOB1-LATS1 NTR domain complex | | Descriptor: | MOB kinase activator 1B, Serine/threonine-protein kinase LATS1, ZINC ION | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
2E4H
 
 | |
2E3I
 
 | |
1UIX
 
 | | Coiled-coil structure of the RhoA-binding domain in Rho-kinase | | Descriptor: | Rho-associated kinase | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel coiled-coil association of the RhoA-binding domain in Rho-kinase
J.Biol.Chem., 278, 2003
|
|
2E3H
 
 | |
5B5V
 
 | | Structure of full-length MOB1b | | Descriptor: | CHLORIDE ION, MOB kinase activator 1B, ZINC ION | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
1WPL
 
 | | Crystal structure of the inhibitory form of rat GTP cyclohydrolase I/GFRP complex | | Descriptor: | 7,8-DIHYDROBIOPTERIN, GTP cyclohydrolase I, GTP cyclohydrolase I feedback regulatory protein, ... | | Authors: | Maita, N, Hatakeyama, K, Okada, K, Hakoshima, T. | | Deposit date: | 2004-09-08 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein
J.Biol.Chem., 279, 2004
|
|
1ISN
 
 | | Crystal structure of merlin FERM domain | | Descriptor: | merlin | | Authors: | Shimizu, T, Seto, A, Maita, N, Hamada, K, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2001-12-13 | | Release date: | 2002-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain.
J.Biol.Chem., 277, 2002
|
|
3A8Q
 
 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3WX1
 
 | | Mouse Cereblon thalidomide binding domain, selenomethionine derivative | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3WX2
 
 | | Mouse Cereblon thalidomide binding domain, native | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3W6X
 
 | | Yeast N-acetyltransferase Mpr1 in complex with CHOP | | Descriptor: | (4S)-4-hydroxy-L-proline, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8N
 
 | | Crystal structure of the Tiam1 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
2ZSH
 
 | | Structural basis of gibberellin(GA3)-induced DELLA recognition by the gibberellin receptor | | Descriptor: | DELLA protein GAI, GIBBERELLIN A3, Probable gibberellin receptor GID1L1 | | Authors: | Murase, K, Hirano, Y, Sun, T.P, Hakoshima, T. | | Deposit date: | 2008-09-10 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gibberellin-induced DELLA recognition by the gibberellin receptor GID1
Nature, 456, 2008
|
|
2E1F
 
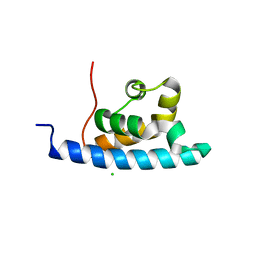 | | Crystal structure of the HRDC Domain of Human Werner Syndrome Protein, WRN | | Descriptor: | CHLORIDE ION, Werner syndrome ATP-dependent helicase | | Authors: | Kitano, K, Yoshihara, N, Hakoshima, T. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the HRDC domain of human Werner syndrome protein, WRN
J.Biol.Chem., 282, 2007
|
|
2EMS
 
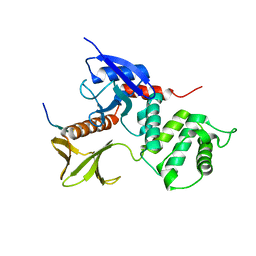 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule CD43 | | Descriptor: | Leukosialin, Radixin | | Authors: | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the cytoplasmic tail of adhesion molecule CD43 and its binding to ERM proteins
J.Mol.Biol., 381, 2008
|
|
3W6S
 
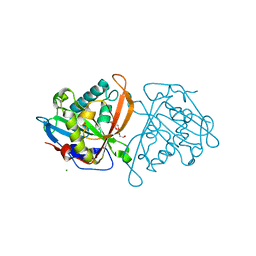 | | yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2EMT
 
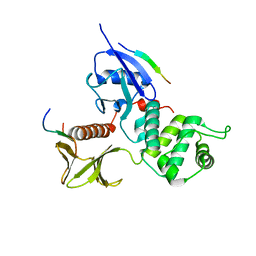 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1 | | Descriptor: | P-selectin glycoprotein ligand 1, Radixin | | Authors: | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of PSGL-1 binding to ERM proteins
Genes Cells, 12, 2007
|
|
2E1E
 
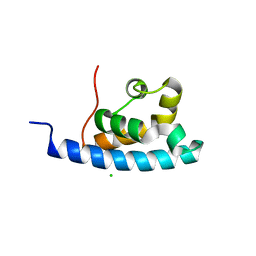 | | Crystal structure of the HRDC Domain of Human Werner Syndrome Protein, WRN | | Descriptor: | CHLORIDE ION, Werner syndrome ATP-dependent helicase | | Authors: | Kitano, K, Yoshihara, N, Hakoshima, T. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the HRDC domain of human Werner syndrome protein, WRN
J.Biol.Chem., 282, 2007
|
|
1VTG
 
 | | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), triostin A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular structure of a DNA-triostin A complex.
Science, 225, 1984
|
|
3W91
 
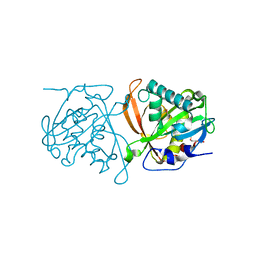 | | crystal structure of SeMet-labeled yeast N-acetyltransferase Mpr1 L87M mutant | | Descriptor: | MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-03-23 | | Release date: | 2013-07-17 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2ZPY
 
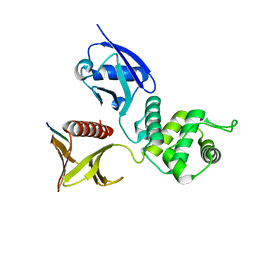 | | Crystal structure of the mouse radxin FERM domain complexed with the mouse CD44 cytoplasmic peptide | | Descriptor: | CD44 antigen, Radixin | | Authors: | Mori, T, Kitano, K, Terawaki, S, Maesaki, R, Fukami, Y, Hakoshima, T. | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for CD44 recognition by ERM proteins
J.Biol.Chem., 283, 2008
|
|
2ZSI
 
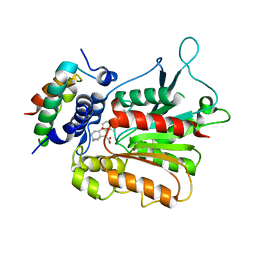 | | Structural basis of gibberellin(GA4)-induced DELLA recognition by the gibberellin receptor | | Descriptor: | DELLA protein GAI, GIBBERELLIN A4, Probable gibberellin receptor GID1L1 | | Authors: | Murase, K, Hirano, Y, Sun, T.P, Hakoshima, T. | | Deposit date: | 2008-09-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gibberellin-induced DELLA recognition by the gibberellin receptor GID1
Nature, 456, 2008
|
|
