6NN3
 
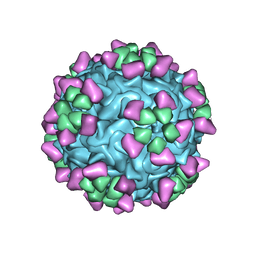 | | Structure of parvovirus B19 decorated with Fab molecules from a human antibody | | Descriptor: | Fab monoclonal antibody 860-55D, heavy chain, light chain, ... | | Authors: | Rossmann, M.G, Sun, Y, Klose, T, Liu, Y. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-13 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of Parvovirus B19 Decorated by Fabs from a Human Antibody.
J. Virol., 93, 2019
|
|
6MWC
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-5 antibody | | Descriptor: | E1, E2, EEEV-5 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6WDT
 
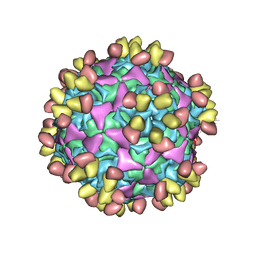 | | Enterovirus D68 in complex with human monoclonal antibody EV68-228 | | Descriptor: | EV68-228 heavy chain, EV68-228 light chain, viral protein 1, ... | | Authors: | Fu, J, Vogt, M.R, Klose, T, Crowe, J.E, Rossmann, M.G, Kuhn, R.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human antibodies neutralize enterovirus D68 and protect against infection and paralytic disease.
Sci Immunol, 5, 2020
|
|
6PSF
 
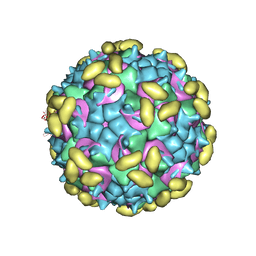 | | Rhinovirus C15 complexed with domains I and II of receptor CDHR3 | | Descriptor: | Cadherin-related family member 3, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7T9P
 
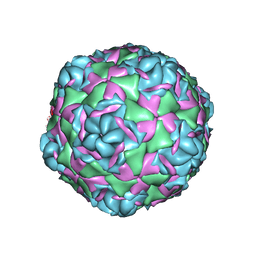 | |
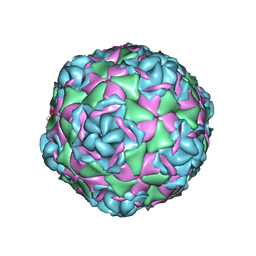
7TAH
 
 | |
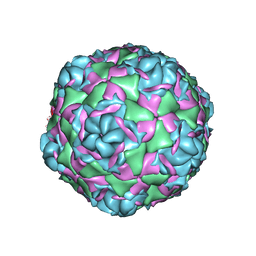
7TAJ
 
 | |
7TAF
 
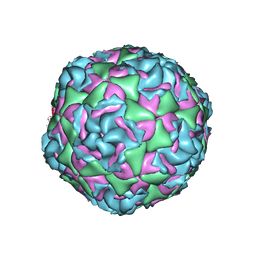 | | Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with inhibitor 11526092 | | Descriptor: | N,N-dimethyl-5-(3-{2-methyl-4-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]phenoxy}propyl)-1,2-oxazole-3-carboxamide, viral protein 1, viral protein 2, ... | | Authors: | Fu, J, Klose, T, Kuhn, R.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Isoxazole-3-Carboxamide Derivatives of Pleconaril Destabilize the Viral Capsid of Enterovirus-D68
To Be Published
|
|
7TAG
 
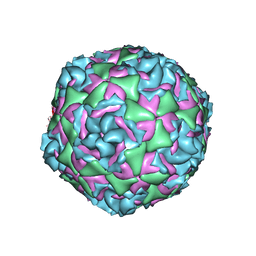 | | Cryo-EM structure of Human Enterovirus D68 US/MO/14-18947 strain virion in complex with pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, viral protein 1, viral protein 2, ... | | Authors: | Fu, J, Klose, T, Kuhn, R.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Isoxazole-3-Carboxamide Derivatives of Pleconaril Destabilize the Viral Capsid of Enterovirus-D68
To Be Published
|
|
7XJB
 
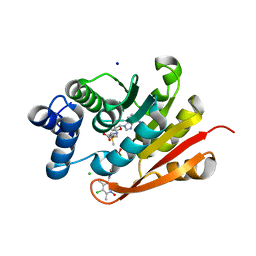 | | Rat-COMT, opicapone,SAM and Mg bond | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Takebe, K, Iijima, H, Suzuki, M, Kuwada-Kusunose, T. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
7XGI
 
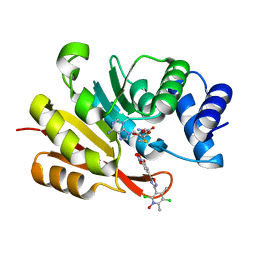 | | COMT SAH Mg opicapone complex | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, Opicapone, ... | | Authors: | Takebe, K, Kuwada-Kusunose, T, Suzuki, M, Iijima, H. | | Deposit date: | 2022-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
7KH1
 
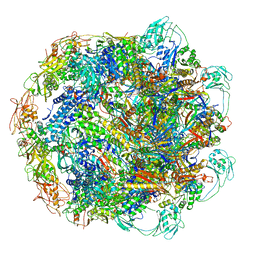 | | Baseplate Complex for Myoviridae Phage XM1 | | Descriptor: | baseplate organization protein, gp11, baseplate stabilizing protein, ... | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
7KLN
 
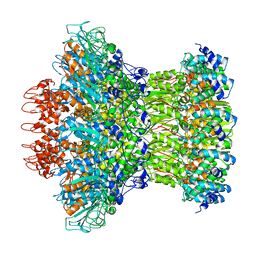 | | Myoviridae Phage XM1 Neck Region (12-fold) | | Descriptor: | Head completion protein, gp1, Portal protein | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-10-30 | | Release date: | 2021-11-03 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
7KJK
 
 | | The Neck region of Phage XM1 (6-fold symmetry) | | Descriptor: | Collar spike protein, Head completion protein, Tail sheath protein, ... | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
7KMX
 
 | | The capsid of Myoviridae Phage XM1 | | Descriptor: | Major capsid protein, Minor capsid protein | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-11-03 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
3GJL
 
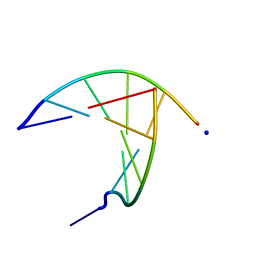 | | crystal structure of a DNA duplex containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3', SODIUM ION | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S, Haraguchi, T, Xiao, M, Kurose, T, Ohkubo, A, Sekine, M, Shibata, T, Millington, C.L, Williams, D.M. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into the stabilizing contributions of bicyclic cytosine analogues: crystal structures of DNA duplexes containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one
To be Published
|
|
6K3N
 
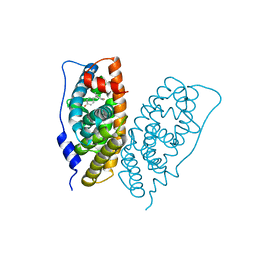 | |
8T0Q
 
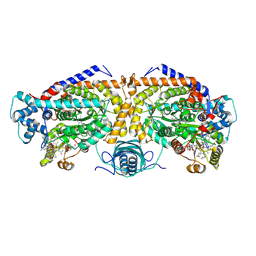 | | Open state of lysine 5,6-aminomutase from Thermoanaerobacter tengcongensis | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-lysine 5,6-aminomutase alpha subunit, ... | | Authors: | Tian, S, Voss, P, Pham, K, Klose, T. | | Deposit date: | 2023-06-01 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Catalysis in Motion: Large-Scale Domain Shift Enables Co-C Bond Homolysis in Lysine 5,6-Aminomutase
To Be Published
|
|
8T0V
 
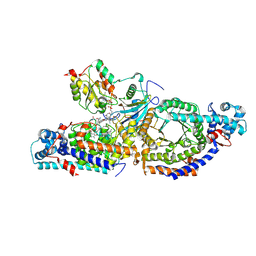 | | Closed state of lysine 5,6-aminomutase from Thermoanaerobacter tengcongensis | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-lysine 5,6-aminomutase alpha subunit, ... | | Authors: | Tian, S, Voss, P, Pham, K, Klose, T. | | Deposit date: | 2023-06-01 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Catalysis in Motion: Large-Scale Domain Shift Enables Co-C Bond Homolysis in Lysine 5,6-Aminomutase
To Be Published
|
|
7N69
 
 | |
7N6A
 
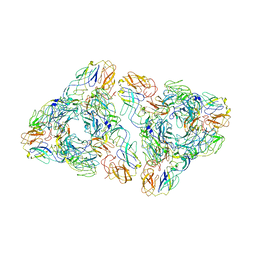 | |
7LCG
 
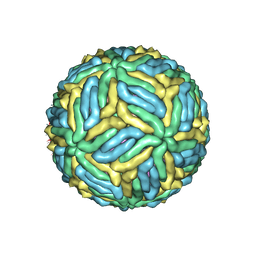 | | The mature Usutu SAAR-1776, Model A | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Khare, B, Klose, T, Fang, Q, Kuhn, R. | | Deposit date: | 2021-01-11 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LCH
 
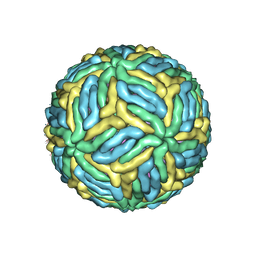 | | The mature Usutu SAAR-1776, Model B | | Descriptor: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Khare, B, Klose, T, Fang, Q, Kuhn, R. | | Deposit date: | 2021-01-11 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7V0N
 
 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21 | | Descriptor: | IgG 21 Fab heavy chain, IgG 21 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7V0O
 
 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94 | | Descriptor: | IgG 94 Fab heavy chain, IgG 94 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
