2DWN
 
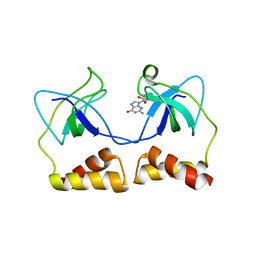 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
4F84
 
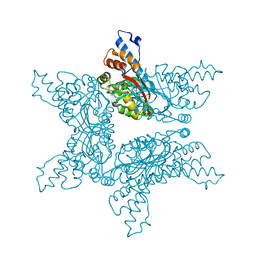 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with SAM | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2DWL
 
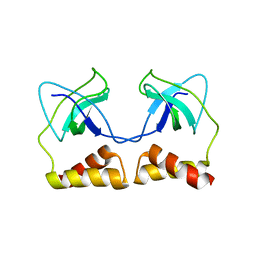 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*(DC))-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
1J0B
 
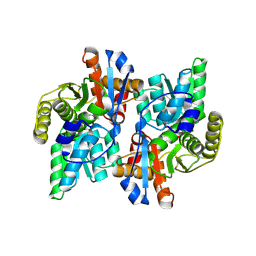 | | Crystal Structure Analysis of the ACC deaminase homologue complexed with inhibitor | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Fujino, A, Ose, T, Honma, M, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and enzymatic properties of 1-aminocyclopropane-1-carboxylate deaminase homologue from Pyrococcus horikoshii
J.Mol.Biol., 341, 2004
|
|
2DWM
 
 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D3V
 
 | | Crystal Structure of Leukocyte Ig-like Receptor A5 (LILRA5/LIR9/ILT11) | | Descriptor: | leukocyte immunoglobulin-like receptor subfamily A member 5 isoform 1 | | Authors: | Shiroishi, M, Kajikawa, M, Kuroki, K, Ose, T, Kohda, D, Maenaka, K. | | Deposit date: | 2005-10-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the human monocyte-activating receptor,
J.Biol.Chem., 281, 2006
|
|
3W39
 
 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
3WV0
 
 | | O-glycan attached to herpes simplex virus type 1 glycoprotein gB is recognized by the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | Envelope glycoprotein B, N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-galactopyranose, Paired immunoglobulin-like type 2 receptor alpha, ... | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WUZ
 
 | | Crystal structure of the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | CITRIC ACID, ISOPROPYL ALCOHOL, Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WO9
 
 | | Crystal structure of the lamprey variable lymphocyte receptor C | | Descriptor: | Variable lymphocyte receptor C | | Authors: | Kanda, R, Sutoh, Y, Kasamatsu, J, Maenaka, K, Kasahara, M, Ose, T. | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the lamprey variable lymphocyte receptor C reveals an unusual feature in its N-terminal capping module.
Plos One, 9, 2014
|
|
5SWZ
 
 | | Crystal Structure of NP1-B17 TCR-H2Db-NP complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Gras, S, Del Campo, C.M, Farenc, C, Josephs, T.M, Rossjohn, J. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Reversed T Cell Receptor Docking on a Major Histocompatibility Class I Complex Limits Involvement in the Immune Response.
Immunity, 45, 2016
|
|
5SWS
 
 | | Crystal Structure of NP2-B17 TCR-H2Db-NP complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Gras, S, Del Campo, C.M, Farenc, C, Josephs, T.M, Rossjohn, J. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Reversed T Cell Receptor Docking on a Major Histocompatibility Class I Complex Limits Involvement in the Immune Response.
Immunity, 45, 2016
|
|
3WY8
 
 | | Crystal Structure of Protease Anisep from Arthrobacter Nicotinovorans | | Descriptor: | Serine protease | | Authors: | Sone, T, Haraguchi, Y, Kuwahara, A, Ose, T, Takano, M, Abe, A, Tanaka, M, Tanaka, I, Asano, K. | | Deposit date: | 2014-08-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization reveals the keratinolytic activity of an arthrobacter nicotinovorans protease.
Protein Pept.Lett., 22, 2015
|
|
3VI6
 
 | | Crystal Structure of human ribosomal protein L30e | | Descriptor: | 60S ribosomal protein L30, FORMIC ACID | | Authors: | Kawaguchi, A, Ose, T, Yao, M, Tanaka, I. | | Deposit date: | 2011-09-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallization and preliminary X-ray structure analysis of human ribosomal protein L30e
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3WBK
 
 | | crystal structure analysis of eukaryotic translation initiation factor 5B and 1A complex | | Descriptor: | Eukaryotic translation initiation factor 1A, Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WH2
 
 | | Human Mincle in complex with citrate | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION, CITRATE ANION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WHD
 
 | | C-type lectin, human MCL | | Descriptor: | C-type lectin domain family 4 member D, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WBI
 
 | | Crystal structure analysis of eukaryotic translation initiation factor 5B structure I | | Descriptor: | Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WMD
 
 | | Crystal structure of epoxide hydrolase MonBI | | Descriptor: | Probable monensin biosynthesis isomerase | | Authors: | Minami, A, Ose, T, Sato, K, Oikawa, A, Kuroki, K, Maenaka, K, Oguri, H, Oikawa, H. | | Deposit date: | 2013-11-16 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Allosteric regulation of epoxide opening cascades by a pair of epoxide hydrolases in monensin biosynthesis
Acs Chem.Biol., 9, 2014
|
|
3WH3
 
 | | human Mincle, ligand free form | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WBJ
 
 | | Crystal structure analysis of eukaryotic translation initiation factor 5B structure II | | Descriptor: | Eukaryotic translation initiation factor 5B | | Authors: | Zheng, A, Yamamoto, R, Ose, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-05-20 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1F2D
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Yao, M, Ose, T, Sugimoto, H, Horiuchi, A, Nakagawa, A, Yokoi, D, Murakami, T, Honma, M, Wakatsuki, S, Tanaka, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
J.Biol.Chem., 275, 2000
|
|
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | Descriptor: | MCPv1, MCPv2, MCPv3, ... | | Authors: | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
5KHF
 
 | |
5K0U
 
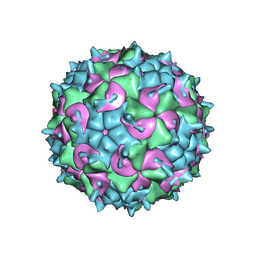 | | CryoEM structure of the full virion of a human rhinovirus C | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Hill, M.G, Klose, T, Chen, Z, Watters, K.E, Jiang, W, Palmenberg, A.C, Rossmann, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
