5XSO
 
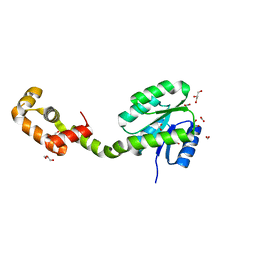 | | Crystal structure of full-length FixJ from B. japonicum crystallized in space group C2221 | | Descriptor: | FORMIC ACID, GLYCEROL, Response regulator FixJ | | Authors: | Nishizono, Y, Hisano, T, Sawai, H, Shiro, Y, Nakamura, H, Wright, G.S.A, Saeki, A, Hikima, T, Yamamoto, M, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system.
Sci Signal, 11, 2018
|
|
5XT2
 
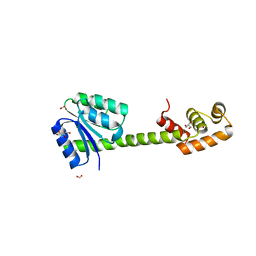 | | Crystal structures of full-length FixJ from B. japonicum crystallized in space group P212121 | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nishizono, Y, Hisano, T, Shiro, Y, Sawai, H, Wright, G.S.A, Saeki, A, Hikima, T, Nakamura, H, Yamamoto, M, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system.
Sci Signal, 11, 2018
|
|
6AIR
 
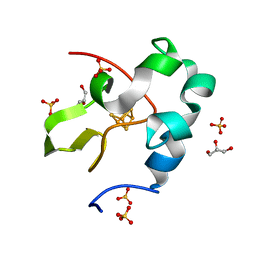 | | High resolution structure of perdeuterated high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
6AIQ
 
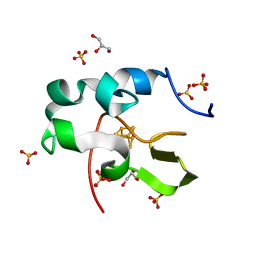 | | High resolution structure of recombinant high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Characterization of perdeuterated high-potential iron-sulfur protein with high-resolution X-ray crystallography.
Proteins, 88, 2020
|
|
5ZCA
 
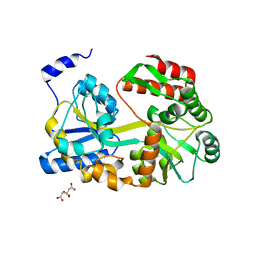 | | Crystal structure of lambda repressor (1-20) fused with maltose-binding protein | | Descriptor: | CITRIC ACID, Repressor protein cI,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2018-02-16 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Co-translational folding of alpha-helical proteins: structural studies of intermediate-length variants of the lambda repressor
Febs Open Bio, 8, 2018
|
|
7VOS
 
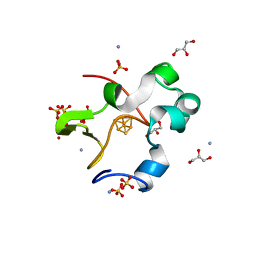 | | High-resolution neutron and X-ray joint refined structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | AMMONIUM ION, GLYCEROL, High-potential iron-sulfur protein, ... | | Authors: | Hanazono, Y, Hirano, Y, Takeda, K, Kusaka, K, Tamada, T, Miki, K. | | Deposit date: | 2021-10-14 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (0.66 Å), X-RAY DIFFRACTION | | Cite: | Revisiting the concept of peptide bond planarity in an iron-sulfur protein by neutron structure analysis.
Sci Adv, 8, 2022
|
|
3VQL
 
 | |
7EDC
 
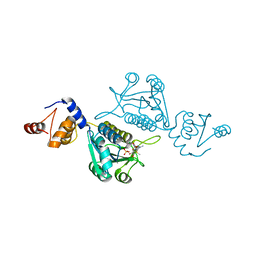 | | Crystal structure of mutant tRNA [Gm18] methyltransferase TrmH (E107G) in complex with S-adenosyl-L-methionine from Escherichia coli | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (guanosine(18)-2'-O)-methyltransferase | | Authors: | Kono, Y, Ito, A, Okamoto, A, Yamagami, R, Hirata, A, Hori, H. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Unique substrate specificity of type II tRNA Gm18 methyltransferase from Escherichia coli
To Be Published
|
|
3WOA
 
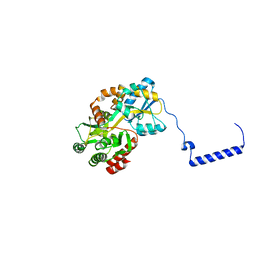 | |
3VQM
 
 | |
5B3Z
 
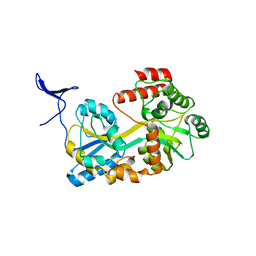 | |
5ZUI
 
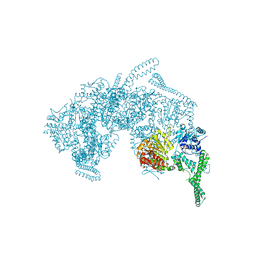 | | Crystal Structure of HSP104 from Chaetomium thermophilum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat Shock Protein 104, SULFATE ION | | Authors: | Hanazono, Y, Inoue, Y, Noguchi, K, Yohda, M, Shinohara, K, Takeda, K, Miki, K. | | Deposit date: | 2018-05-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 2021
|
|
5B3P
 
 | |
5BMY
 
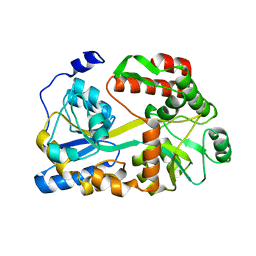 | |
5B3X
 
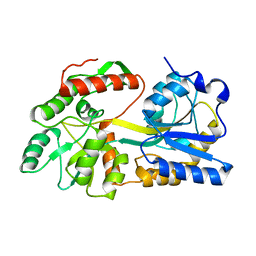 | |
5CL2
 
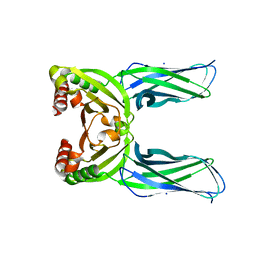 | | Crystal structure of Spo0M, sporulation control protein, from Bacillus subtilis. | | Descriptor: | SODIUM ION, Sporulation-control protein spo0M | | Authors: | Sonoda, Y, Mizutani, K, Mikami, B. | | Deposit date: | 2015-07-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Spo0M, a sporulation-control protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
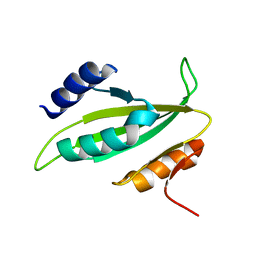
5B3Q
 
 | |
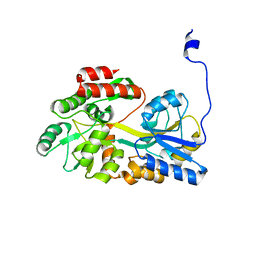
5B3Y
 
 | |
5B3W
 
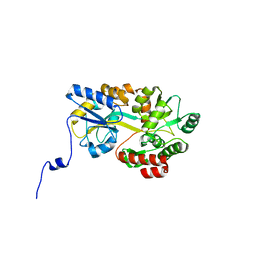 | | Crystal structure of hPin1 WW domain (5-15) fused with maltose-binding protein in C2221 form | | Descriptor: | CITRIC ACID, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2016-03-17 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of the N-terminal fragments of the WW domain: Insights into co-translational folding of a beta-sheet protein
Sci Rep, 6, 2016
|
|
2M1W
 
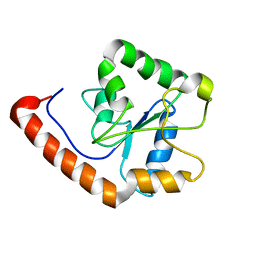 | | TICAM-2 TIR domain | | Descriptor: | TIR domain-containing adapter molecule 2 | | Authors: | Enokizono, Y, Kumeta, H, Funami, K, Horiuchi, M, Sarmiento, J, Yamashita, K, Standley, D.M, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M1X
 
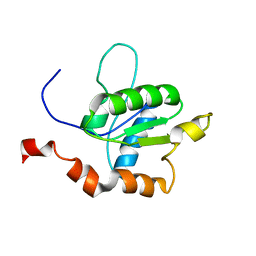 | | TICAM-1 TIR domain structure | | Descriptor: | TIR domain-containing adapter molecule 1 | | Authors: | Enokizono, Y, Kumeta, H, Funami, K, Horiuchi, M, Sarmiento, J, Yamashita, K, Standley, D.M, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VQK
 
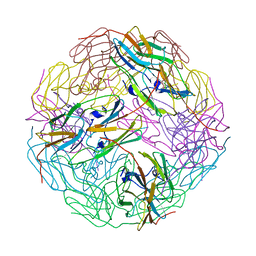 | |
3W1Z
 
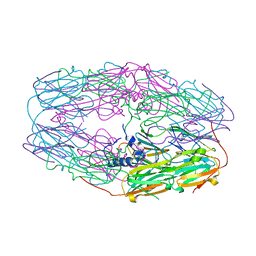 | | Heat shock protein 16.0 from Schizosaccharomyces pombe | | Descriptor: | Heat shock protein 16 | | Authors: | Hanazono, Y, Takeda, K, Akiyama, N, Aikawa, Y, Miki, K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Nonequivalence Observed for the 16-Meric Structure of a Small Heat Shock Protein, SpHsp16.0, from Schizosaccharomyces pombe
Structure, 21, 2013
|
|
2DQM
 
 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | Authors: | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-29 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
1WTB
 
 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D (AUF1) with telomere DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2004-11-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D
J.Biol.Chem., 280, 2005
|
|
