7E5O
 
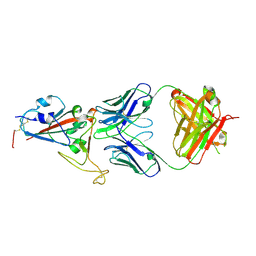 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
7PBJ
 
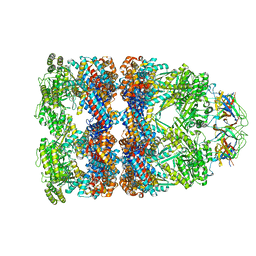 | |
7PBX
 
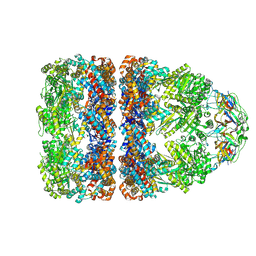 | |
4Q4U
 
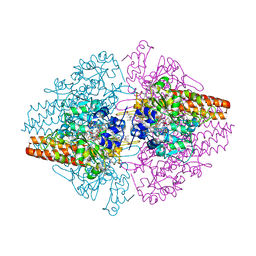 | | TvNiR in complex with sulfite, low dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
to be published
|
|
4Q17
 
 | | Free form of TvNiR, middle dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-03 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
To be published
|
|
4Q0T
 
 | | Free form of TvNiR, low dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-02 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
TO BE PUBLISHED
|
|
4Q5B
 
 | | TvNiR in complex with sulfite, high dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
To be Published
|
|
8Q9X
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum with molecular oxygen at 1.05 A resolution | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-08-22 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
8Q9Y
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum in complex with inhibitor thiourea at 1.10 A resolution | | Descriptor: | COPPER (II) ION, GLYCEROL, THIOUREA, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2023-08-22 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum at atomic resolution
To Be Published
|
|
4Q1O
 
 | | Free form of TvNiR, high dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-04 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
To be Published
|
|
4Q5C
 
 | | TvNiR in complex with sulfite, middle dose data set | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Lazarenko, V.A, Polyakov, K.M, Trofimov, A.A, Popov, A.N, Tikhonova, T.V, Tikhonov, A.V, Popov, V.O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-09-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray-induced changes in the active site structure of octaheme cytochrome c nitrite reductase and its substrate complexes
To be Published
|
|
1V94
 
 | | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix | | Descriptor: | isocitrate dehydrogenase | | Authors: | Jeong, J.-J, Sonoda, T, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2004-01-20 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix
Proteins, 55, 2004
|
|
5GW7
 
 | |
1UG9
 
 | | Crystal Structure of Glucodextranase from Arthrobacter globiformis I42 | | Descriptor: | CALCIUM ION, GLYCEROL, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Ohtaki, A, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1T08
 
 | | Crystal structure of beta-catenin/ICAT helical domain/unphosphorylated APC R3 | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin, Beta-catenin-interacting protein 1 | | Authors: | Ha, N.-C, Tonozuka, T, Stamos, J.L, Weis, W.I. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Mol.Cell, 15, 2004
|
|
5UUO
 
 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase-like protein, ... | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
4CPH
 
 | | trans-divalent streptavidin with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
5UUN
 
 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
4CPF
 
 | | Wild-type streptavidin in complex with love-hate ligand 3 (LH3) | | Descriptor: | STREPTAVIDIN, methyl 4-(2-{5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H- thieno[3,4-d]imidazolidin-4-yl]pentanehydrazido}-3- [4-(methoxycarbonyl)phenyl]phenyl)benzoate | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
1UH3
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase/acarbose complex | | Descriptor: | (1S,2S,3R,6R)-6-amino-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Abe, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2003-06-23 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complex Structures of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 with Malto-oligosaccharides Demonstrate the Role of Domain N Acting as a Starch-binding Domain
J.Mol.Biol., 335, 2004
|
|
1UH4
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1/malto-tridecaose complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2003-06-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 with Malto-oligosaccharides Demonstrate the Role of Domain N Acting as a Starch-binding Domain
J.Mol.Biol., 335, 2004
|
|
4CPI
 
 | | streptavidin A86D mutant with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
1VB9
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) complexed with transglycosylated product | | Descriptor: | CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-[alpha-D-glucopyranose-(1-6)]alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-amylase II | | Authors: | Mizuno, M, Tonozuka, T, Uechi, A, Ohtaki, A, Ichikawa, K, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-02-25 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) complexed with transglycosylated product
EUR.J.BIOCHEM., 271, 2004
|
|
6I3Q
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus complex with acetate ions. | | Descriptor: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Polyakov, K.M, Popov, A.N, Tikhkonova, T.V, Popov, V.O, Trofimov, A.A. | | Deposit date: | 2018-11-07 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4CPE
 
 | | Wild-type streptavidin in complex with love-hate ligand 1 (LH1) | | Descriptor: | (3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N-{2-[(2,6- diphenylphenyl)formamido]ethyl}pentanamide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
