7QE1
 
 | | Crystal structure of apo SN243 | | Descriptor: | SN243, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEA
 
 | | Crystal structure of fluorescein-di-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{S})-3,4,5-tris(oxidanyl)-6-[(1~{R})-6'-oxidanyl-3-oxidanylidene-spiro[2-benzofuran-1,9'-xanthene]-3'-yl]oxy-oxane-2-carboxylic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QG4
 
 | | Apo crystal structure of a mutant of SN243 (D415N) | | Descriptor: | SN243, SULFATE ION, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEF
 
 | | Crystal structure of para-nitrophenyl-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
8CJV
 
 | |
8CI3
 
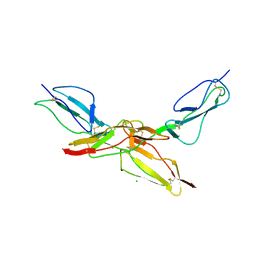 | | Structure of bovine CD46 ectodomain (SCR 1-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Aitkenhead, H, David I Stuart, D.I, El Omari, K. | | Deposit date: | 2023-02-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of Bovine CD46 Ectodomain.
Viruses, 15, 2023
|
|
8CKM
 
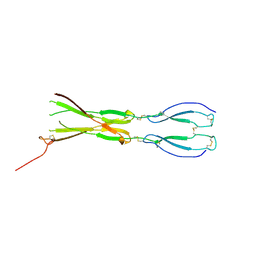 | | Semaphorin-5A TSR 3-4 domains | | Descriptor: | Semaphorin-5A | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
8CKG
 
 | | Semaphorin-5A TSR 3-4 domains in complex with sulfate | | Descriptor: | SULFATE ION, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
8CKK
 
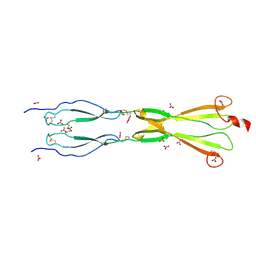 | | Semaphorin-5A TSR 3-4 domains in complex with nitrate | | Descriptor: | NITRATE ION, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
8CKL
 
 | | Semaphorin-5A TSR 3-4 domains in complex with sucrose octasulfate (SOS) | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-2)-1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose, Semaphorin-5A, alpha-D-mannopyranose | | Authors: | Nagy, G.N, Duman, R, Harlos, K, El Omari, K, Wagner, A, Jones, E.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure and function of Semaphorin-5A glycosaminoglycan interactions.
Nat Commun, 15, 2024
|
|
6OWX
 
 | | Spy H96L:Im7 L18pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWY
 
 | | Spy H96L:Im7 K20pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2J41
 
 | | Crystal structure of Staphylococcus aureus guanylate monophosphate kinase | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANYLATE KINASE, POTASSIUM ION, ... | | Authors: | El Omari, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Staphylococcus Aureus Guanylate Monophosphate Kinase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2J0F
 
 | | Structural basis for non-competitive product inhibition in human thymidine phosphorylase: implication for drug design | | Descriptor: | THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | El Omari, K, Bronckaers, A, Liekens, S, Perez-Perez, M.J, Balzarini, J, Stammers, D.K. | | Deposit date: | 2006-08-02 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Non-Competitive Product Inhibition in Human Thymidine Phosphorylase: Implications for Drug Design.
Biochem.J., 399, 2006
|
|
2J87
 
 | | Structure of vaccinia virus thymidine kinase in complex with dTTP: insights for drug design | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | El Omari, K, Solaroli, N, Karlsson, A, Balzarini, J, Stammers, D.K. | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Vaccinia Virus Thymidine Kinase in Complex with Dttp: Insights for Drug Design.
Bmc Struct.Biol., 6, 2006
|
|
6OWZ
 
 | | Spy H96L:Im7 L19pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7OGS
 
 | |
7OOT
 
 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element | | Descriptor: | DNA (5'-D(P*AP*GP*CP*TP*TP*TP*CP*TP*CP*GP*GP*TP*TP*TP*CP*AP*GP*TP*TP*G)-3'), DNA (5'-D(P*TP*CP*AP*AP*CP*TP*GP*AP*AP*AP*CP*CP*GP*AP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor 4, ... | | Authors: | Agnarelli, A, El Omari, K, Alt, A.O, Mancini, E.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to interferon-stimulated response element
To Be Published
|
|
7PGM
 
 | | HHIP-C in complex with heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-3)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
7PGK
 
 | | HHIP-N, the N-terminal domain of the Hedgehog-Interacting Protein (HHIP), in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
7PGL
 
 | | HHIP-N, the N-terminal domain of the Hedgehog-Interacting Protein (HHIP), apo-form | | Descriptor: | Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
7PGN
 
 | | HHP-C in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
6JJH
 
 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJF
 
 | | Crystal structure of a two-quartet DNA mixed-parallel/antiparallel G-quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*CP*TP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3'), POTASSIUM ION, ... | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
5MDU
 
 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe. | | Descriptor: | CHLORIDE ION, GLYCEROL, Rpb7-binding protein seb1, ... | | Authors: | Wittmann, S, Renner, M, El Omari, K, Adams, O, Vasiljeva, L, Grimes, J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA.
Nat Commun, 8, 2017
|
|
