1G6K
 
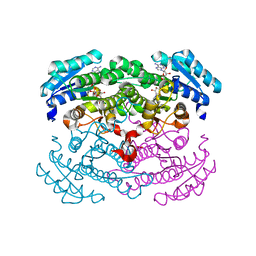 | | Crystal structure of glucose dehydrogenase mutant E96A complexed with NAD+ | | 分子名称: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | 登録日 | 2000-11-06 | | 公開日 | 2003-08-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
1GCO
 
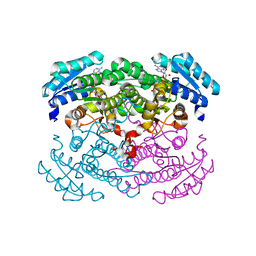 | | CRYSTAL STRUCTURE OF GLUCOSE DEHYDROGENASE COMPLEXED WITH NAD+ | | 分子名称: | GLUCOSE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | 登録日 | 2000-08-07 | | 公開日 | 2001-02-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of glucose dehydrogenase from Bacillus megaterium IWG3 at 1.7 A resolution.
J.Biochem., 129, 2001
|
|
6OGP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-063 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, 1,2-ETHANEDIOL, Protease | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGT
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-001 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGL
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-003 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-02 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGV
 
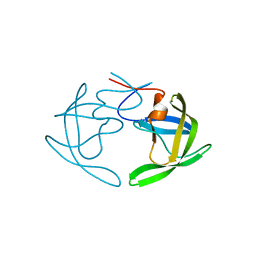 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in apo state | | 分子名称: | Protease | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
6OGQ
 
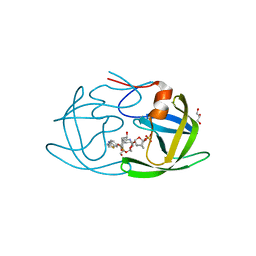 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-003 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGS
 
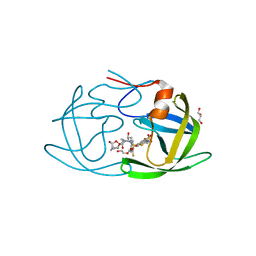 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-001 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OYD
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-004 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-05-14 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGR
 
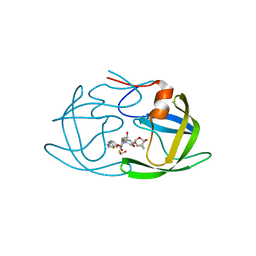 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-142 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
6OYR
 
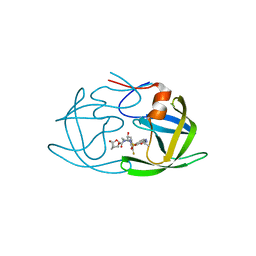 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-002 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-05-15 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
1GAW
 
 | |
2E1U
 
 | |
2E1V
 
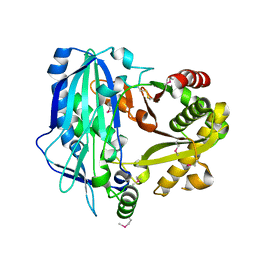 | | Crystal structure of Dendranthema morifolium DmAT, seleno-methionine derivative | | 分子名称: | acyl transferase | | 著者 | Unno, H, Ichimaida, F, Kusunoki, M, Nakayama, T. | | 登録日 | 2006-10-28 | | 公開日 | 2007-04-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and Mutational Studies of Anthocyanin Malonyltransferases Establish the Features of BAHD Enzyme Catalysis
J.Biol.Chem., 282, 2007
|
|
6ND6
 
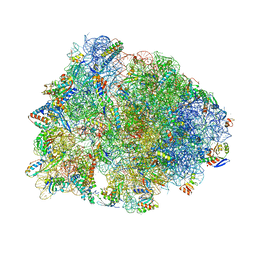 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with erythromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.85A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | 登録日 | 2018-12-13 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
6ND5
 
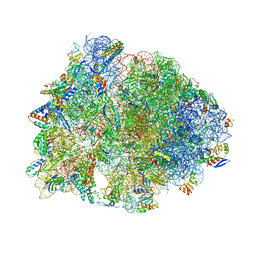 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with chloramphenicol and bound to mRNA and A-, P-, and E-site tRNAs at 2.60A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | 登録日 | 2018-12-13 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
8YGJ
 
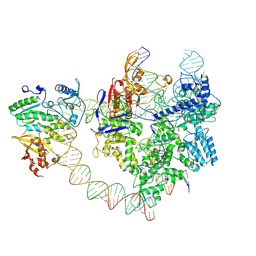 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2024-02-26 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUU
 
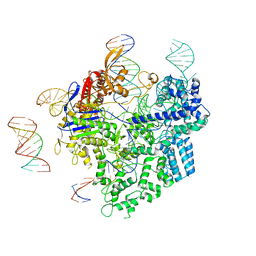 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUV
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUT
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
7QLR
 
 | |
6MCS
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-003 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | 著者 | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-09-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
6MCR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-001 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | 著者 | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-09-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
1VDY
 
 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | 分子名称: | hypothetical protein (RAFL09-17-B18) | | 著者 | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-03-25 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
