2YX9
 
 | | Crystal structure of D298K copper amine oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2007-04-25 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Trapping of a dopaquinone intermediate in the TPQ cofactor biogenesis in a copper-containing amine oxidase from Arthrobacter globiformis.
J.Am.Chem.Soc., 129, 2007
|
|
1UKJ
 
 | | Detailed structure of L-Methionine-Lyase from Pseudomonas putida | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2003-08-24 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detailed structure of L-Methionine -Lyase from Pseudomonas putida
To be Published
|
|
2PED
 
 | | Crystallographic model of 9-cis-rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Nakamichi, H, Okada, T. | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Photoisomerization mechanism of rhodopsin and 9-cis-rhodopsin revealed by x-ray crystallography
Biophys.J., 92, 2007
|
|
3AMO
 
 | | Time-resolved X-ray Crystal Structure Analysis of Enzymatic Reaction of Copper Amine Oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Kataoka, M, Oya, H, Tominaga, A, Otsu, M, Okajima, T, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2010-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detection of the reaction intermediates catalyzed by a copper amine oxidase.
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3UON
 
 | | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, CHLORIDE ION, Human M2 muscarinic acetylcholine, ... | | Authors: | Haga, K, Kruse, A.C, Asada, H, Yurugi-Kobayashi, T, Shiroishi, M, Zhang, C, Weis, W.I, Okada, T, Kobilka, B.K, Haga, T, Kobayashi, T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist.
Nature, 482, 2012
|
|
3VOC
 
 | | Crystal structure of the catalytic domain of beta-amylase from paenibacillus polymyxa | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta/alpha-amylase, ... | | Authors: | Nishimura, S, Fujioka, T, Nakaniwa, T, Tada, T. | | Deposit date: | 2012-01-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis by X-ray crystallography and small-angle scattering of the multi-domain beta-amylase from Paenibacillus polymyxa
To be Published
|
|
3WA3
 
 | | Crystal structure of copper amine oxidase from arthrobacter globiformis in N2 condition | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WA2
 
 | | High resolution crystal structure of copper amine oxidase from arthrobacter globiformis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1CD9
 
 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
2GLT
 
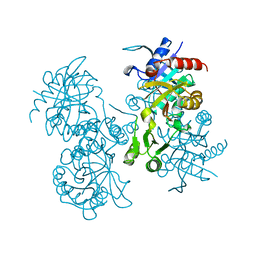 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
2CH8
 
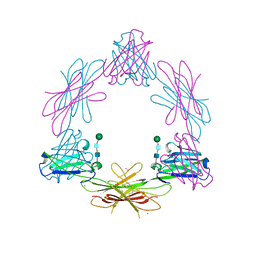 | | Structure of the Epstein-Barr Virus Oncogene BARF1 | | Descriptor: | 33 KDA EARLY PROTEIN, PLATINUM (II) ION, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Tarbouriech, N, Ruggiero, F, deTurenne-Tessier, M, Ooka, T, Burmeister, W.P. | | Deposit date: | 2006-03-13 | | Release date: | 2006-05-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Epstein-Barr Virus Oncogene Barf1
J.Mol.Biol., 359, 2006
|
|
3BIS
 
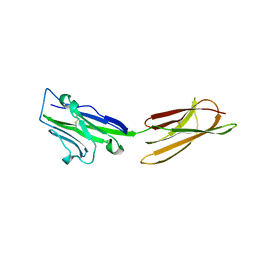 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1HZX
 
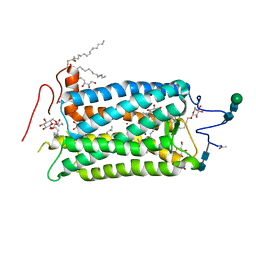 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Teller, D.C, Okada, T, Behnke, C.A, Palczewski, K, Stenkamp, R.E. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs).
Biochemistry, 40, 2001
|
|
3BIK
 
 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3VZV
 
 | | Crystal structure of human mdm2 with a dihydroimidazothiazole inhibitor | | Descriptor: | 1-{[(5R,6S)-5,6-bis(4-chlorophenyl)-6-methyl-3-(propan-2-yl)-5,6-dihydroimidazo[2,1-b][1,3]thiazol-2-yl]carbonyl}-N,N-dimethyl-L-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shimizu, H, Katakura, S, Miyazaki, M, Naito, H, Sugimoto, Y, Kawato, H, Okayama, T, Soga, T. | | Deposit date: | 2012-10-16 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lead optimization of novel p53-MDM2 interaction inhibitors possessing dihydroimidazothiazole scaffold
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3W69
 
 | | Crystal structure of human mdm2 with a dihydroimidazothiazole inhibitor | | Descriptor: | (5R,6S)-2-[((2S,5R)-2-{[(3R)-4-acetyl-3-methylpiperazin-1-yl]carbonyl}-5-ethylpyrrolidin-1-yl)carbonyl]-5,6-bis(4-chlorophenyl)-3-isopropyl-6-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shimizu, H, Katakura, S, Miyazaki, M, Naito, H, Sugimoto, Y, Kawato, H, Okayama, T, Soga, T. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of novel orally active p53-MDM2 interaction inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
3WKV
 
 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | Descriptor: | Ion channel | | Authors: | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.453 Å) | | Cite: | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3VTZ
 
 | |
1HLV
 
 | | CRYSTAL STRUCTURE OF CENP-B(1-129) COMPLEXED WITH THE CENP-B BOX DNA | | Descriptor: | CENP-B BOX DNA, MAJOR CENTROMERE AUTOANTIGEN B | | Authors: | Tanaka, Y, Nureki, O, Kurumizaka, H, Fukai, S, Kawaguchi, S, Ikuta, M, Iwahara, J, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-12-04 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA.
EMBO J., 20, 2001
|
|
1IQX
 
 | | CRYSTAL STRUCTURE OF COBALT-SUBSTITUTED AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | Descriptor: | CO(II)-SUBSTITUTED AMINE OXIDASE, COBALT (II) ION | | Authors: | Kishishita, S, Okajima, T, Mure, M, Kim, M, Yamaguchi, H, Hirota, S, Suzuki, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2001-08-27 | | Release date: | 2003-02-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
1IVU
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IQY
 
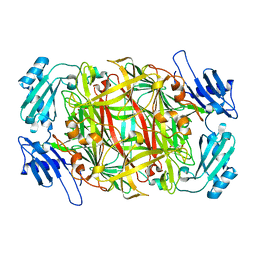 | | CRYSTAL STRUCTURE OF NICKEL-SUBSTITUTED AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | Descriptor: | AMINE OXIDASE, NICKEL (II) ION | | Authors: | Kishishita, S, Okajima, T, Mure, M, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2001-08-28 | | Release date: | 2003-02-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
1IVV
 
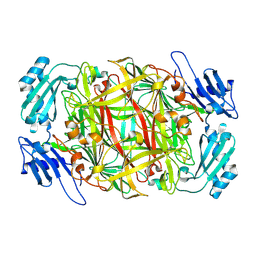 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Early intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IU7
 
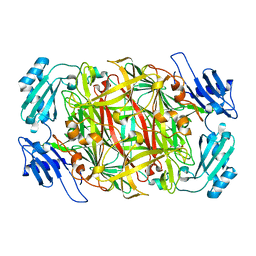 | | HOLO FORM OF COPPER-CONTAINING AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS | | Descriptor: | AMINE OXIDASE, COPPER (II) ION | | Authors: | Kishishita, S, Okajima, T, Kim, M, Yamaguchi, H, Hirota, S, Suzuki, S, Kuroda, S, Tanizawa, K, Mure, M. | | Deposit date: | 2002-02-28 | | Release date: | 2003-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Copper Ion in Bacterial Copper Amine Oxidase: Spectroscopic and Crystallographic Studies of Metal-Substituted Enzymes
J.AM.CHEM.SOC., 125, 2003
|
|
1IVW
 
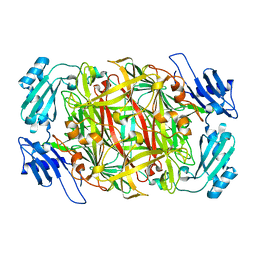 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Late intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
