4PK5
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with Amg-1 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(1,3-benzodioxol-5-yl)-2-{[5-(4-methylphenyl)[1,3]thiazolo[2,3-c][1,2,4]triazol-3-yl]sulfanyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kohno, T, Tojo, S, Ishii, T. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structures and Structure-Activity Relationships of Imidazothiazole Derivatives as IDO1 Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PK6
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with imidazothiazole derivative | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-[2-(3-chlorophenyl)ethyl]-3-(4-methylphenyl)imidazo[2,1-b][1,3]thiazole-5-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kohno, T, Tojo, S, Ishii, T, Kamioka, S. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal Structures and Structure-Activity Relationships of Imidazothiazole Derivatives as IDO1 Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
1OMG
 
 | | NMR STUDY OF OMEGA-CONOTOXIN MVIIA | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Kohno, T, Kim, J.-I, Kobayashi, K, Kodera, Y, Maeda, T, Sato, K. | | Deposit date: | 1995-04-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the calcium channel blocker omega-conotoxin MVIIA.
Biochemistry, 34, 1995
|
|
1FU3
 
 | |
1V5A
 
 | |
1QCM
 
 | | AMYLOID BETA PEPTIDE (25-35), NMR, 20 STRUCTURES | | Descriptor: | AMYLOID BETA PEPTIDE | | Authors: | Kohno, T, Kobayashi, K, Maeda, T, Sato, K, Takashima, A. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structures of the amyloid beta peptide (25-35) in membrane-mimicking environment.
Biochemistry, 35, 1996
|
|
2D1N
 
 | | Collagenase-3 (MMP-13) complexed to a hydroxamic acid inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, SM-25453, ... | | Authors: | Kohno, T, Hochigai, H, Yamashita, E, Tsukihara, T, Kanaoka, M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structures of the catalytic domain of human stromelysin-1 (MMP-3) and collagenase-3 (MMP-13) with a hydroxamic acid inhibitor SM-25453
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2D1O
 
 | | Stromelysin-1 (MMP-3) complexed to a hydroxamic acid inhibitor | | Descriptor: | CALCIUM ION, SM-25453, Stromelysin-1, ... | | Authors: | Kohno, T, Hochigai, H, Yamashita, E, Tsukihara, T, Kanaoka, M. | | Deposit date: | 2005-08-30 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of the catalytic domain of human stromelysin-1 (MMP-3) and collagenase-3 (MMP-13) with a hydroxamic acid inhibitor SM-25453
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2D52
 
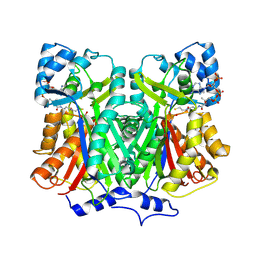 | |
2D51
 
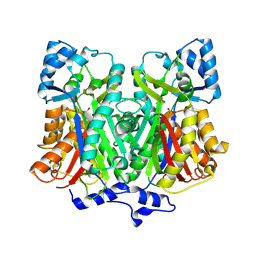 | | Pentaketide chromone synthase (M207G mutant) | | Descriptor: | pentaketide chromone synthase | | Authors: | Kohno, T, Morita, H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insight into Chain-Length Control and Product Specificity of Pentaketide Chromone Synthase from Aloe arborescens
Chem.Biol., 14, 2007
|
|
3VPH
 
 | | L-lactate dehydrogenase from Thermus caldophilus GK24 complexed with oxamate, NADH and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Arai, K, Ohno, T, Miyanaga, A, Fushinobu, S, Taguchi, H. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The core of allosteric motion in Thermus caldophilus L-lactate dehydrogenase.
J.Biol.Chem., 2014
|
|
3VPG
 
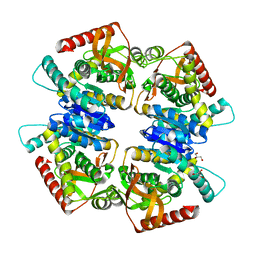 | | L-lactate dehydrogenase from Thermus caldophilus GK24 | | Descriptor: | GLYCEROL, L-lactate dehydrogenase | | Authors: | Arai, K, Ohno, T, Miyanaga, A, Fushinobu, S, Taguchi, H. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The core of allosteric motion in Thermus caldophilus L-lactate dehydrogenase.
J.Biol.Chem., 2014
|
|
7EN4
 
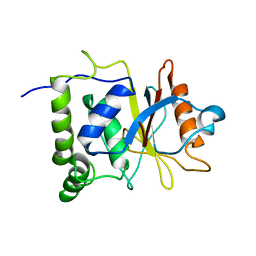 | | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase YUH1 | | Authors: | Okada, M, Tateishi, Y, Nojiri, E, Mikawa, T, Rajesh, S, Ogasawa, H, Ueda, T, Yagi, H, Kohno, T, Kigawa, T, Shimada, I, Guentert, P, Yutaka, I, Ikeya, T. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase.
To Be Published
|
|
6FEK
 
 | |
1A13
 
 | | G PROTEIN-BOUND CONFORMATION OF MASTOPARAN-X, NMR, 14 STRUCTURES | | Descriptor: | MASTOPARAN-X | | Authors: | Kusunoki, H, Wakamatsu, K, Sato, K, Miyazawa, T, Kohno, T. | | Deposit date: | 1997-12-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | G protein-bound conformation of mastoparan-X: heteronuclear multidimensional transferred nuclear overhauser effect analysis of peptide uniformly enriched with 13C and 15N.
Biochemistry, 37, 1998
|
|
2RRN
 
 | | Solution structure of SecDF periplasmic domain P4 | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tanaka, T, Tsukazaki, T, Echizen, Y, Nureki, O, Kohno, T. | | Deposit date: | 2011-01-30 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2MLQ
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2RQ5
 
 | |
1NC8
 
 | | HIGH-RESOLUTION SOLUTION NMR STRUCTURE OF THE MINIMAL ACTIVE DOMAIN OF THE HUMAN IMMUNODEFICIENCY VIRUS TYPE-2 NUCLEOCAPSID PROTEIN, 15 STRUCTURES | | Descriptor: | NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Kodera, Y, Sato, K, Tsukahara, T, Komatsu, H, Maeda, T, Kohno, T. | | Deposit date: | 1998-05-14 | | Release date: | 1999-05-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution NMR structure of the minimal active domain of the human immunodeficiency virus type-2 nucleocapsid protein.
Biochemistry, 37, 1998
|
|
2RQ1
 
 | |
4ZKF
 
 | | Crystal structure of human phosphodiesterase 12 | | Descriptor: | 2',5'-phosphodiesterase 12, MAGNESIUM ION | | Authors: | Kim, S.Y, Kohno, T, Mori, T, Kitano, K, Hakoshima, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human phosphodiesterase
To Be Published
|
|
1J1C
 
 | | Binary complex structure of human tau protein kinase I with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase kinase-3 beta, MAGNESIUM ION | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J1B
 
 | | Binary complex structure of human tau protein kinase I with AMPPNP | | Descriptor: | Glycogen synthase kinase-3 beta, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1F3K
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF OMEGA-CONOTOXIN TXVII, AN L-TYPE CALCIUM CHANNEL BLOCKER | | Descriptor: | OMEGA-CONOTOXIN TXVII | | Authors: | Kobayashi, K, Sasaki, T, Sato, K, Kohno, T. | | Deposit date: | 2000-06-05 | | Release date: | 2000-12-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of omega-conotoxin TxVII, an L-type calcium channel blocker.
Biochemistry, 39, 2000
|
|
