7XGG
 
 | |
7XGF
 
 | |
7XGE
 
 | |
1POO
 
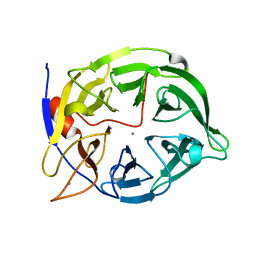 | | THERMOSTABLE PHYTASE FROM BACILLUS SP | | Descriptor: | CALCIUM ION, PROTEIN (PHYTASE) | | Authors: | Oh, B.H, Ha, N.C. | | Deposit date: | 1999-04-16 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1AYP
 
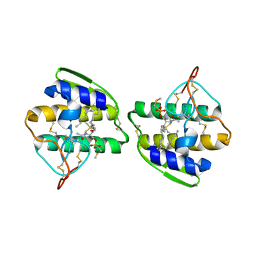 | |
3BL2
 
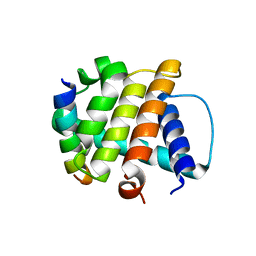 | | Crystal Structure of M11, the BCL-2 Homolog of Murine Gamma-herpesvirus 68, Complexed with Mouse Beclin1 (residues 106-124) | | Descriptor: | Beclin-1, V-bcl-2 | | Authors: | Oh, B.-H, Woo, J.-S, Ku, B. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral BCL-2 of Murine gamma-Herpesvirus 68
Plos Pathog., 4, 2008
|
|
1E9Y
 
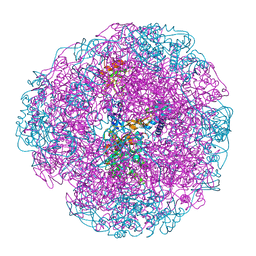 | | Crystal structure of Helicobacter pylori urease in complex with acetohydroxamic acid | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, UREASE SUBUNIT ALPHA, ... | | Authors: | Ha, N.-C, Oh, S.-T, Oh, B.-H. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease
Nat.Struct.Biol., 8, 2001
|
|
1W6Y
 
 | | crystal structure of a mutant W92A in ketosteroid isomerase (KSI) from Pseudomonas putida biotype B | | Descriptor: | BETA-MERCAPTOETHANOL, EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Yun, Y.S, Nam, G.H, Kim, Y.-G, Oh, B.-H, Choi, K.Y. | | Deposit date: | 2004-08-25 | | Release date: | 2005-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Exterior Hydrophobic Cluster Contributes to Conformational Stability and Steroid Binding in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
FEBS J., 272, 2005
|
|
1E97
 
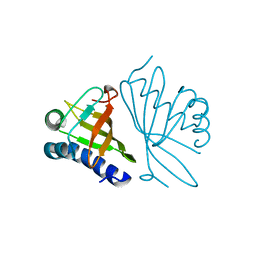 | |
1EA9
 
 | | Cyclomaltodextrinase | | Descriptor: | CYCLOMALTODEXTRINASE | | Authors: | Cho, H.-S, Kim, M.-S, Oh, B.-H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cyclomaltodextrinase, Neopullulanase, and Maltogenic Amylase are Nearly Indistinguishable from Each Other
J.Biol.Chem., 277, 2002
|
|
5X1E
 
 | |
7DG5
 
 | |
4UW2
 
 | | Crystal structure of Csm1 in T.onnurineus | | Descriptor: | CSM1 | | Authors: | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-25 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
4YJ4
 
 | | Crystal structure of Bcl-xL in complex with the BIM BH3 domain containing Ile155-to-Arg and Glu158-to-phosphoserine mutations | | Descriptor: | ACETATE ION, Bcl-2-like protein 1, BIM BH3 domain, ... | | Authors: | Ku, B, Ha, N.-C, Oh, B.-H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-29 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of cell-survival activity of Akt into apoptotic death of cancer cells by two mutations on the BIM BH3 domain.
Cell Death Dis, 6, 2015
|
|
2POO
 
 | | THERMOSTABLE PHYTASE IN FULLY CALCIUM LOADED STATE | | Descriptor: | CALCIUM ION, PROTEIN (PHYTASE) | | Authors: | Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-04-16 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
8CHO
 
 | |
7VYT
 
 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | Descriptor: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | Authors: | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
1H6L
 
 | |
5GKX
 
 | | Crystal structure of TON_0340, apo form | | Descriptor: | PHOSPHATE ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL2
 
 | | Crystal structure of TON_0340 in complex with Ca | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL3
 
 | | Crystal structure of TON_0340 in complex with Mg | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL4
 
 | | Crystal structure of TON_0340 in complex with Mn | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
7VYR
 
 | |
7CBC
 
 | |
