1V8J
 
 | | The Crystal Structure of the Minimal Functional Domain of the Microtubule Destabilizer KIF2C Complexed with Mg-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2C, MAGNESIUM ION | | Authors: | Ogawa, T, Nitta, R, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops.
Cell(Cambridge,Mass.), 116, 2004
|
|
1V8K
 
 | | The Crystal Structure of the Minimal Functional Domain of the Microtubule Destabilizer KIF2C Complexed with Mg-AMPPNP | | Descriptor: | Kinesin-like protein KIF2C, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ogawa, T, Nitta, R, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops.
Cell(Cambridge,Mass.), 116, 2004
|
|
7EO9
 
 | | Crystal structure of KIF1A Motor-Neck domain with ADP-Mg-AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Kinesin-like protein KIF1A, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A neuropathy-associated kinesin KIF1A mutation hyper-stabilizes the motor-neck interaction during the ATPase cycle.
Embo J., 41, 2022
|
|
7EOB
 
 | | Crystal structure of KIF1A Motor-Neck domain E239K mutant with ADP-Mg-AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Kinesin-like protein KIF1A, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A neuropathy-associated kinesin KIF1A mutation hyper-stabilizes the motor-neck interaction during the ATPase cycle.
Embo J., 41, 2022
|
|
5XJB
 
 | | The Crystal Structure of the Minimal Core Domain of the Microtubule Depolymerizer KIF2C Complexed with ADP-Mg-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Kinesin-like protein KIF2C, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2017-04-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation
Cell Rep, 20, 2017
|
|
5XJA
 
 | | The Crystal Structure of the Minimal Core Domain of the Microtubule Depolymerizer KIF2C Complexed with ADP-Mg-AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Kinesin-like protein KIF2C, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2017-04-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation
Cell Rep, 20, 2017
|
|
8W7P
 
 | | Extracellular domain of a sensor histidine kinase | | Descriptor: | Extracellular domain of a sensor histidine kinase NagS, GLYCEROL | | Authors: | Itoh, T, Ogawa, T, Hibi, T, Kimoto, H. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of the extracellular domain of sensor histidine kinase NagS from Paenibacillus sp. str. FPU-7: nagS interacts with oligosaccharide binding protein NagB1 in complexes with N, N'-diacetylchitobiose.
Biosci.Biotechnol.Biochem., 88, 2024
|
|
1C1F
 
 | | LIGAND-FREE CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I) | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1C1L
 
 | | LACTOSE-LIGANDED CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I), beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
2DFX
 
 | | Crystal structure of the carboxy terminal domain of colicin E5 complexed with its inhibitor | | Descriptor: | Colicin-E5, Colicin-E5 immunity protein | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-03-06 | | Release date: | 2007-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
2DJH
 
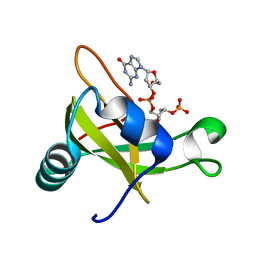 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-04-03 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
1WLC
 
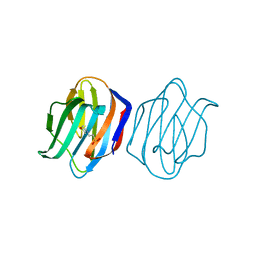 | | Congerin II Y16S/T88I double mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Congerin II | | Authors: | Shionyu-Mitsuyama, C, Ito, Y, Konno, A, Miwa, Y, Ogawa, T, Muramoto, K, Shirai, T. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro evolutionary thermostabilization of congerin II: a limited reproduction of natural protein evolution by artificial selection pressure
J.Mol.Biol., 347, 2005
|
|
1WLD
 
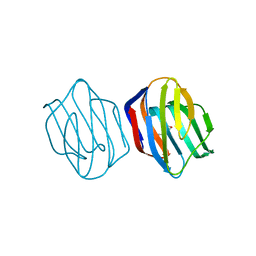 | | Congerin II T88I single mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shionyu-Mitsuyama, C, Ito, Y, Konno, A, Miwa, Y, Ogawa, T, Muramoto, K, Shirai, T. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In vitro evolutionary thermostabilization of congerin II: a limited reproduction of natural protein evolution by artificial selection pressure
J.Mol.Biol., 347, 2005
|
|
1WLW
 
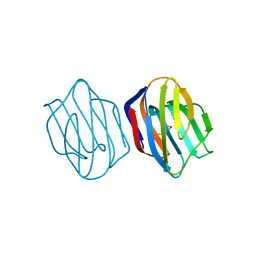 | | Congerin II Y16S single mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Congerin II, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Shionyu-Mitsuyama, C, Ito, Y, Konno, A, Miwa, Y, Ogawa, T, Muramoto, K, Shirai, T. | | Deposit date: | 2004-06-30 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In vitro evolutionary thermostabilization of congerin II: a limited reproduction of natural protein evolution by artificial selection pressure
J.Mol.Biol., 347, 2005
|
|
1TFK
 
 | | Ribonuclease from Escherichia coli complexed with its inhibtor protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
1TFO
 
 | | Ribonuclease from Escherichia coli complexed with its inhibitor protein | | Descriptor: | Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
6JVB
 
 | |
6JV9
 
 | | Crystal Structure of Human CRMP2 1-532, unmodified | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydropyrimidinase-related protein 2, ... | | Authors: | Jiang, X, Ogawa, T, Hirokawa, N. | | Deposit date: | 2019-04-16 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Enhanced carbonyl stress induces irreversible multimerization of CRMP2 in schizophrenia pathogenesis.
Life Sci Alliance, 2, 2019
|
|
1IS4
 
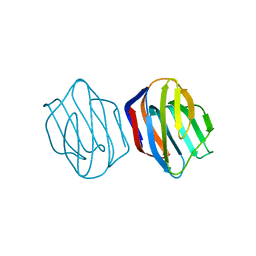 | | LACTOSE-LIGANDED CONGERIN II | | Descriptor: | CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Conger Eel Galectin (Congerin II) at 1.45 A Resolution: Implication for the Accelerated Evolution of a New Ligand-Binding Site Following Gene Duplication
J.Mol.Biol., 321, 2002
|
|
1IS3
 
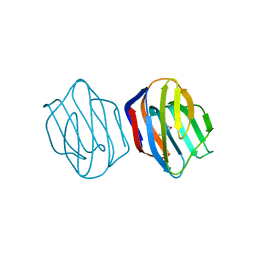 | | LACTOSE AND MES-LIGANDED CONGERIN II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS5
 
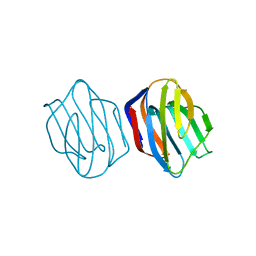 | | Ligand free Congerin II | | Descriptor: | Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS6
 
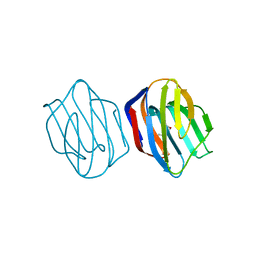 | | MES-Liganded Congerin II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
5ZNM
 
 | | Colicin D Central Domain and C-terminal tRNase domain | | Descriptor: | Colicin-D, GLYCEROL, SULFATE ION | | Authors: | Chang, J.W, Sato, Y, Ogawa, T, Arakawa, T, Fukai, S, Fushinobu, S, Masaki, H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the central and the C-terminal RNase domains of colicin D implicated its translocation pathway through inner membrane of target cell
J. Biochem., 164, 2018
|
|
2ZX0
 
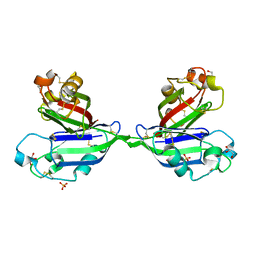 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, GLYCEROL, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX2
 
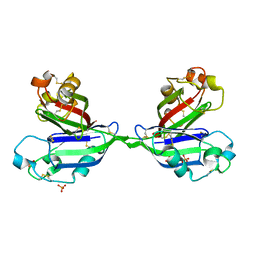 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-L-rhamnopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
