7F1L
 
 | | Designed enzyme RA61 M48K/I72D mutant: form V | | 分子名称: | CHLORIDE ION, Engineered Retroaldolase, IMIDAZOLE | | 著者 | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | 登録日 | 2021-06-09 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7CEF
 
 | | Crystal structure of PET-degrading cutinase Cut190 /S226P/R228S/ mutant with the C-terminal three residues deletion | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, ZINC ION | | 著者 | Senga, A, Numoto, N, Ito, N, Kawai, F, Oda, M. | | 登録日 | 2020-06-23 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
J.Biochem., 169, 2021
|
|
7CEH
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant with the C-terminal three residues deletion in ligand ejecting form | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION | | 著者 | Senga, A, Numoto, N, Ito, N, Kawai, F, Oda, M. | | 登録日 | 2020-06-23 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
J.Biochem., 169, 2021
|
|
8IBL
 
 | | MES bound form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | 著者 | Emori, M, Numoto, N, Kamiya, N, Oda, M. | | 登録日 | 2023-02-10 | | 公開日 | 2023-03-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Improvement of thermostability and activity of PET-degrading enzyme Cut190 towards a detailed understanding and application of the enzymatic reaction mechanism.
Biorxiv, 2023
|
|
8IBM
 
 | | Sulfate bound form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, SULFATE ION | | 著者 | Emori, M, Numoto, N, Kamiya, N, Oda, M. | | 登録日 | 2023-02-10 | | 公開日 | 2023-03-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Improvement of thermostability and activity of PET-degrading enzyme Cut190 towards a detailed understanding and application of the enzymatic reaction mechanism.
Biorxiv, 2023
|
|
5GJI
 
 | | PI3K p85 N-terminal SH2 domain/CD28-derived peptide complex | | 分子名称: | GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION, ... | | 著者 | Inaba, S, Numoto, N, Morii, H, Ogawa, S, Ikura, T, Abe, R, Ito, N, Oda, M. | | 登録日 | 2016-06-30 | | 公開日 | 2016-12-14 | | 最終更新日 | 2017-05-10 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
5GJH
 
 | | Gads SH2 domain/CD28-derived peptide complex | | 分子名称: | GRB2-related adapter protein 2, T-cell-specific surface glycoprotein CD28 | | 著者 | Inaba, S, Numoto, N, Morii, H, Ogawa, S, Ikura, T, Abe, R, Ito, N, Oda, M. | | 登録日 | 2016-06-30 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
3BUZ
 
 | | Crystal structure of ia-bTAD-actin complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Tsuge, H, Nagahama, M, Oda, M, Iwamoto, S, Utsunomiya, H, Marquez, V.E, Katunuma, N, Nishizawa, M, Sakurai, J. | | 登録日 | 2008-01-04 | | 公開日 | 2008-05-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5ZRS
 
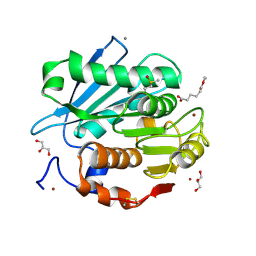 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl adipate bound state | | 分子名称: | 6-ethoxy-6-oxohexanoic acid, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZRR
 
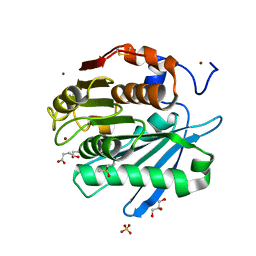 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl succinate bound state | | 分子名称: | 4-ethoxy-4-oxobutanoic acid, Alpha/beta hydrolase family protein, GLYCEROL, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZRQ
 
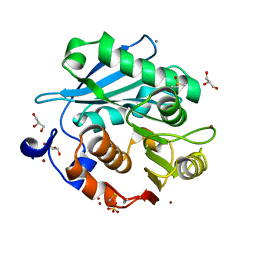 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in Zn(2+)-bound state | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZNO
 
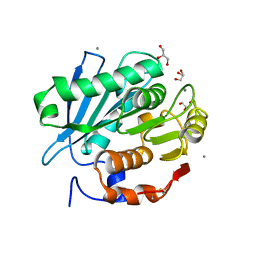 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant in Ca(2+)-bound state | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL | | 著者 | Numoto, N, Inaba, S, Yamagami, Y, Kamiya, N, Bekker, G.J, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-10 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.60264349 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5AUL
 
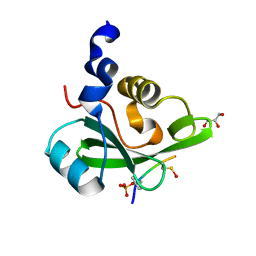 | | PI3K p85 C-terminal SH2 domain/CD28-derived peptide complex | | 分子名称: | GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, T-cell-specific surface glycoprotein CD28 | | 著者 | Inaba, S, Numoto, N, Morii, H, Ikura, T, Oda, M, Ito, N. | | 登録日 | 2015-04-28 | | 公開日 | 2016-05-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
1IDZ
 
 | |
1IDY
 
 | |
6K4Z
 
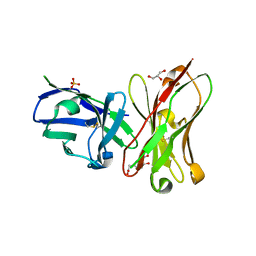 | | Single-chain Fv antibody of C6 COMPLEXED WITH 2-(4-HYDROXY-3-NITROPHENYL)ACETIC ACID | | 分子名称: | 2-(4-HYDROXY-3-NITROPHENYL)ACETIC ACID, GLYCEROL, SULFATE ION, ... | | 著者 | Nishiguchi, A, Numoto, N, Ito, N, Azuma, T, Oda, M. | | 登録日 | 2019-05-27 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Three-dimensional structure of a high affinity anti-(4-hydroxy-3-nitrophenyl)acetyl antibody possessing a glycine residue at position 95 of the heavy chain.
Mol.Immunol., 114, 2019
|
|
3ATG
 
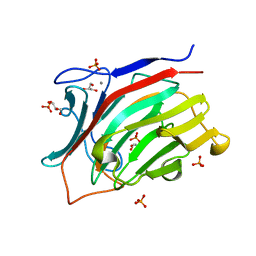 | | endo-1,3-beta-glucanase from Cellulosimicrobium cellulans | | 分子名称: | CALCIUM ION, GLUCANASE, GLYCEROL, ... | | 著者 | Tanabe, Y, Pang, Z, Oda, M, Mikami, B. | | 登録日 | 2011-01-04 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural and thermodynamic characterization of endo-1,3-beta-glucanase: Insights into the substrate recognition mechanism.
Biochim. Biophys. Acta, 1866, 2018
|
|
3WA4
 
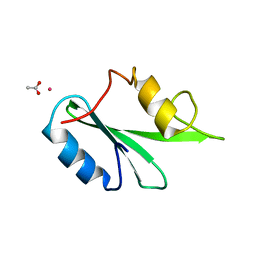 | | Grb2 SH2 domain/CD28-derived peptide complex | | 分子名称: | ACETIC ACID, CADMIUM ION, Growth factor receptor-bound protein 2, ... | | 著者 | Higo, K, Oda, M, Ito, N. | | 登録日 | 2013-04-23 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | High Resolution Crystal Structure of the Grb2 SH2 Domain with a Phosphopeptide Derived from CD28
Plos One, 8, 2013
|
|
2DDS
 
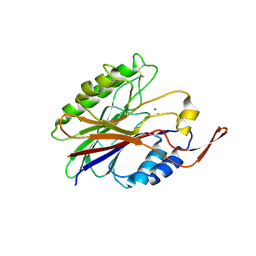 | | Crystal structure of sphingomyelinase from Bacillus cereus with cobalt ion | | 分子名称: | COBALT (II) ION, Sphingomyelin phosphodiesterase | | 著者 | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-02-02 | | 公開日 | 2006-05-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2DDR
 
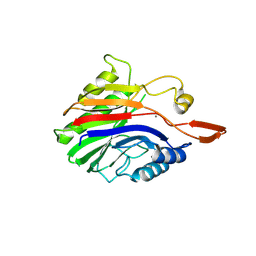 | | Crystal structure of sphingomyelinase from Bacillus cereus with calcium ion | | 分子名称: | CALCIUM ION, Sphingomyelin phosphodiesterase | | 著者 | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-02-02 | | 公開日 | 2006-05-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2DDT
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with magnesium ion | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Ago, H, Oda, M, Tsuge, H, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-02-02 | | 公開日 | 2006-05-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
5JQP
 
 | | Crystal structure of ER glucosidase II heterodimeric complex consisting of catalytic subunit and the binding domain of regulatory subunit | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CALCIUM ION, ... | | 著者 | Satoh, T, Toshimori, T, Noda, M, Uchiyama, S, Kato, K. | | 登録日 | 2016-05-05 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Interaction mode between catalytic and regulatory subunits in glucosidase II involved in ER glycoprotein quality control.
Protein Sci., 25, 2016
|
|
4IJ0
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | 分子名称: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | 著者 | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | 登録日 | 2012-12-20 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
3X26
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 5 min | | 分子名称: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | 著者 | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | 登録日 | 2014-12-10 | | 公開日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X25
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 700 min | | 分子名称: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | 著者 | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | 登録日 | 2014-12-10 | | 公開日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
