3VX6
 
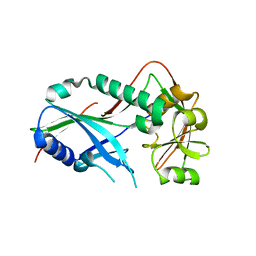 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2E5B
 
 | |
2E5D
 
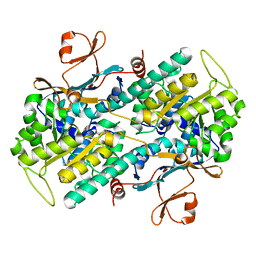
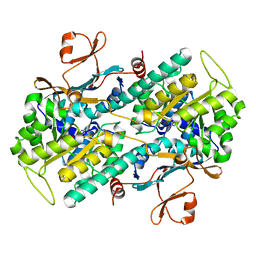 | | Crystal structure of Human NMPRTase complexed with nicotinamide | | Descriptor: | NICOTINAMIDE, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2E5C
 
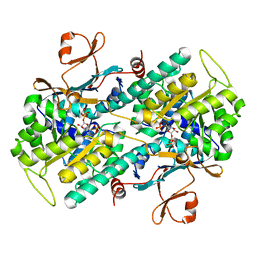 | | Crystal structure of Human NMPRTase complexed with 5'-phosphoribosyl-1'-pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2PA2
 
 | | Crystal structure of human Ribosomal protein L10 core domain | | Descriptor: | 60S ribosomal protein L10, POTASSIUM ION | | Authors: | Nishimura, M, Kaminishi, T, Takemoto, C, Kawazoe, M, Yoshida, T, Tanaka, A, Sugano, S, Shirouzu, M, Ohkubo, T, Yokoyama, S, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Ribosomal Protein L10 Core Domain Reveals Eukaryote-Specific Motifs in Addition to the Conserved Fold
J.Mol.Biol., 377, 2008
|
|
2D0S
 
 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | Descriptor: | HEME C, cytochrome c | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-08-08 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
1IY5
 
 | | Solution structure of wild type OMSVP3 | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | Descriptor: | Collagen-like peptide | | Authors: | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
1T7H
 
 | | X-ray structure of [Lys(-2)-Arg(-1)-des(17-21)]-endothelin-1 peptide | | Descriptor: | Endothelin-1 | | Authors: | Hoh, F, Cerdan, R, Kaas, Q, Nishi, Y, Chiche, L, Kubo, S, Chino, N, Kobayashi, Y, Dumas, C, Aumelas, A. | | Deposit date: | 2004-05-10 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-resolution X-ray structure of the unexpectedly stable dimer of the [Lys(-2)-Arg(-1)-des(17-21)]endothelin-1 peptide
Biochemistry, 43, 2004
|
|
1WKI
 
 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | Descriptor: | LSU ribosomal protein L16P | | Authors: | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
1Y69
 
 | | RRF domain I in complex with the 50S ribosomal subunit from Deinococcus radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, 50S ribosomal protein L27, ... | | Authors: | Wilson, D.N, Schluenzen, F, Harms, J.M, Yoshida, T, Ohkubo, T, Albrecht, R, Buerger, J, Kobayashi, Y, Fucini, P. | | Deposit date: | 2004-12-04 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | X-ray crystallography on ribosome recycling: mechanism of binding and action of RRF on the 50S ribosomal subunit
EMBO J., 24, 2005
|
|
2A4X
 
 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Metal-Free Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
2A4W
 
 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Copper(II)-Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, COPPER (II) ION, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
3VVL
 
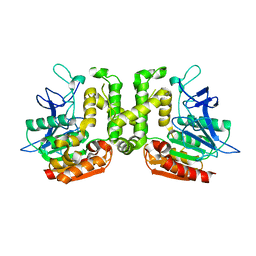 | | Crystal structure of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | Descriptor: | Homoserine O-acetyltransferase | | Authors: | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
2RPV
 
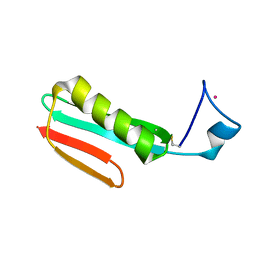 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
2ZW4
 
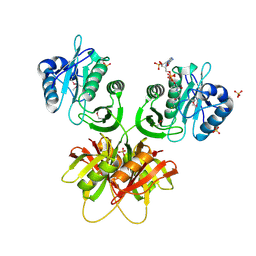 | |
3VVM
 
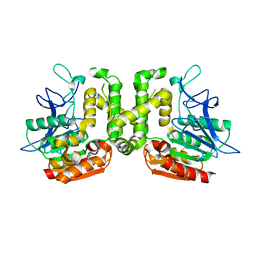 | | Crystal structure of G52A-P55G mutant of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | Descriptor: | Homoserine O-acetyltransferase | | Authors: | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
1VA2
 
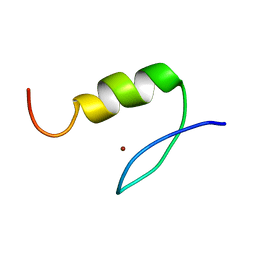 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 2) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
2ZW5
 
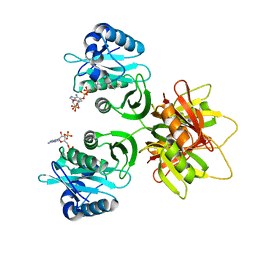 | |
2ZW7
 
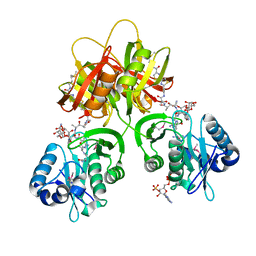 | |
1O5P
 
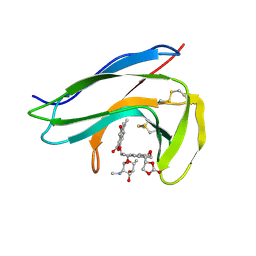 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
1VA3
 
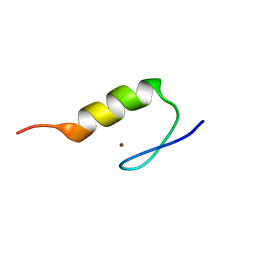 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 3) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
1VA1
 
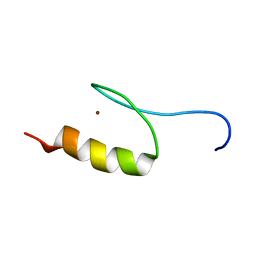 | | Solution Structure of Transcription Factor Sp1 DNA Binding Domain (Zinc Finger 1) | | Descriptor: | Transcription factor Sp1, ZINC ION | | Authors: | Oka, S, Shiraishi, Y, Yoshida, T, Ohkubo, T, Sugiura, Y, Kobayashi, Y. | | Deposit date: | 2004-02-07 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of transcription factor Sp1 DNA binding domain
Biochemistry, 43, 2004
|
|
2ZV6
 
 | | Crystal structure of human squamous cell carcinoma antigen 1 | | Descriptor: | Serpin B3 | | Authors: | Zheng, B, Matoba, Y, Katagiri, C, Hibino, T, Sugiyama, M. | | Deposit date: | 2008-11-01 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SCCA1 and insight about the interaction with JNK1
Biochem.Biophys.Res.Commun., 380, 2009
|
|
