4S13
 
 | | Ferulic Acid Decarboxylase (FDC1) | | Descriptor: | 4-ethenylphenol, Ferulic acid decarboxylase 1 | | Authors: | Lee, S.G, Bhuiya, M.W, Yu, O, Jez, J.M. | | Deposit date: | 2015-01-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Structure and Mechanism of Ferulic Acid Decarboxylase (FDC1) from Saccharomyces cerevisiae.
Appl.Environ.Microbiol., 81, 2015
|
|
1R7A
 
 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | Authors: | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | Deposit date: | 2003-10-21 | | Release date: | 2004-02-10 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
4TKU
 
 | | Reactivated Nitrogenase MoFe-protein from A. vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Spatzal, T, Perez, K, Einsle, O, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-05-27 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase.
Science, 345, 2014
|
|
7QA1
 
 | | The structure of natural crystals of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated using serial femtosecond crystallography at an X-ray free electron laser | | Descriptor: | Toxin-10 pesticidal protein (Tpp) 49Aa1 | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R, Best, H.L. | | Deposit date: | 2021-11-15 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7QS6
 
 | |
7RNT
 
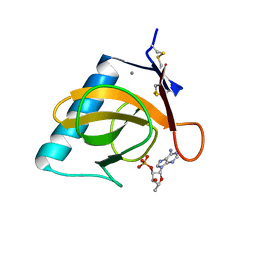 | | CRYSTAL STRUCTURE OF THE TYR45TRP MUTANT OF RIBONUCLEASE T1 IN A COMPLEX WITH 2'-ADENYLIC ACID | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Koellner, G, Grunert, H.-P, Landt, O, Saenger, W. | | Deposit date: | 1991-08-20 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tyr45Trp mutant of ribonuclease T1 in a complex with 2'-adenylic acid.
Eur.J.Biochem., 201, 1991
|
|
4TKV
 
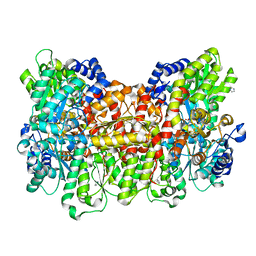 | | CO-bound Nitrogenase MoFe-protein from A. vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CARBON MONOXIDE, FE (II) ION, ... | | Authors: | Spatzal, T, Perez, K, Einsle, O, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase.
Science, 345, 2014
|
|
1RGL
 
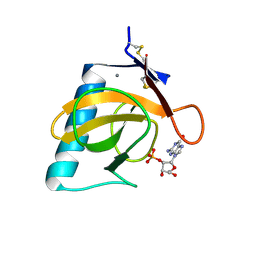 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
7RJJ
 
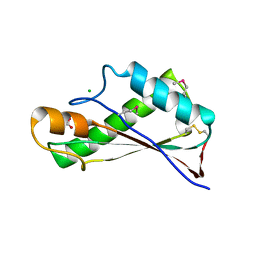 | | Crystal Structure of the Peptidoglycan Binding Domain of the Outer Membrane Protein (OmpA) from Klebsiella pneumoniae with bound D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, OmpA family protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
4RMI
 
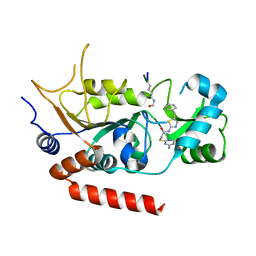 | | Human Sirt2 in complex with SirReal1 and Ac-Lys-OTC peptide | | Descriptor: | Ac-Lys-OTC peptide, N-(5-benzyl-1,3-thiazol-2-yl)-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
1RGK
 
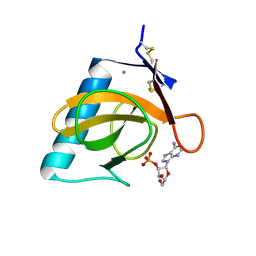 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
1R4D
 
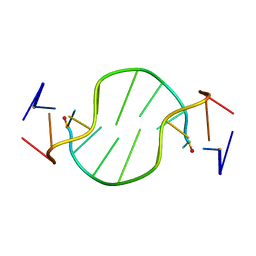 | | Solution structure of the chimeric L/D DNA oligonucleotide d(C8metGCGC(L)G(L)CGCG)2 | | Descriptor: | 5'-D(*CP*(8MG)P*CP*GP*(0DC)P*(0DG)P*CP*GP*CP*G)-3' | | Authors: | Cherrak, I, Mauffret, O, Santamaria, F, Rayner, B, Hocquet, A, Ghomi, M, Fermandjian, S. | | Deposit date: | 2003-10-06 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | L-nucleotides and 8-methylguanine of d(C1m8G2C3G4C5LG6LC7G8C9G10)2 act cooperatively to promote a left-handed helix under physiological salt conditions.
Nucleic Acids Res., 31, 2003
|
|
4RPC
 
 | | Crystal structure of the putative alpha/beta hydrolase family protein from Desulfitobacterium hafniense | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TETRAETHYLENE GLYCOL, putative alpha/beta hydrolase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the putative alpha/beta hydrolase family protein from Desulfitobacterium hafniense
To be Published
|
|
1RU3
 
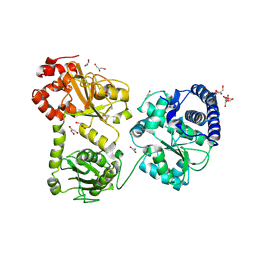 | | Crystal Structure of the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans | | Descriptor: | Acetyl-CoA synthase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Svetlitchnyi, V, Dobbek, H, Meyer-Klaucke, W, Meins, T, Thiele, B, Rmer, P, Huber, R, Meyer, O. | | Deposit date: | 2003-12-11 | | Release date: | 2003-12-23 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A functional Ni-Ni-[4Fe-4S] cluster in the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4S31
 
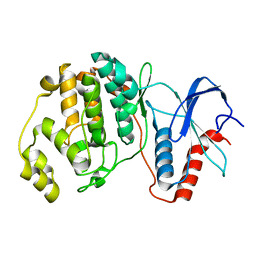 | |
1R8G
 
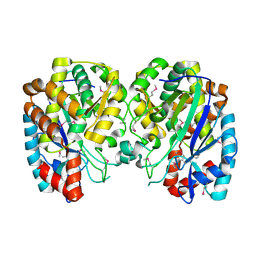 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1RLW
 
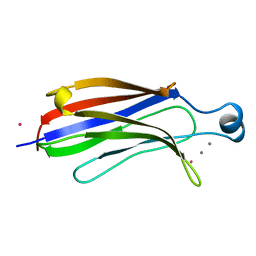 | |
1RFR
 
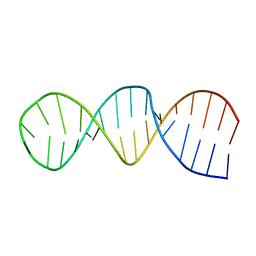 | | NMR structure of the 30mer stemloop-D of coxsackieviral RNA | | Descriptor: | stemloop-D RNA of the 5'-cloverleaf of coxsackievirus B3 | | Authors: | Ohlenschlager, O, Wohnert, J, Bucci, E, Seitz, S, Hafner, S, Ramachandran, R, Zell, R, Gorlach, M. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf
RNA and its interaction with the proteinase 3C.
STRUCTURE, 12, 2004
|
|
7RB6
 
 | | Low temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
7RB7
 
 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA and MMB4 oxime | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
7RB5
 
 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
4RMJ
 
 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RSH
 
 | | Structure of a putative lipolytic protein of G-D-S-L family from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CHLORIDE ION, Lipolytic protein G-D-S-L family | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-07 | | Release date: | 2014-11-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a putative lipolytic protein of G-D-S-L family from Desulfitobacterium hafniense DCB-2
To be Published
|
|
4RMH
 
 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
7RDJ
 
 | | Crystal structure of PCDN-16B, an anti-HIV antibody from the PCDN bnAb lineage (non-cysteinylated state) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PCDN-16B Fab heavy chain, ... | | Authors: | Omorodion, O, Wilson, I.A. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Biochemical Characterization of Cysteinylation in Broadly Neutralizing Antibodies to HIV-1.
J.Mol.Biol., 433, 2021
|
|
