1ZYK
 
 | | Anthranilate Phosphoribosyltransferase in complex with PRPP, anthranilate and magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase, ... | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
2FLH
 
 | | Crystal structure of cytokinin-specific binding protein from mung bean in complex with cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, SODIUM ION, cytokinin-specific binding protein | | Authors: | Pasternak, O, Bujacz, G.D, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Vigna radiata Cytokinin-Specific Binding Protein in Complex with Zeatin.
Plant Cell, 18, 2006
|
|
1BPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 9), INSULIN B CHAIN (PH 9), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
5TR3
 
 | | 2.5 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD.
To Be Published
|
|
5TRE
 
 | | Zinc and the Iron Donor Frataxin Regulate Oligomerization of the Scaffold Protein to Form New Fe-S Cluster Assembly Centers | | Descriptor: | Frataxin homolog, mitochondrial, Iron sulfur cluster assembly protein 1 | | Authors: | Ranatunga, W, Gakh, O, Galeano, B.K, Smith IV, D.Y, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-10-26 | | Release date: | 2017-06-07 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (15.6 Å) | | Cite: | Zinc and the iron donor frataxin regulate oligomerization of the scaffold protein to form new Fe-S cluster assembly centers.
Metallomics, 9, 2017
|
|
7KZX
 
 | | Cryo-EM structure of YiiP-Fab complex in Apo state | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2R heavy chain, Fab2R light chain | | Authors: | Lopez-Redondo, M.L, Fan, S, Koide, A, Koide, S, Beckstein, O, Stokes, D.L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-06 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP.
J.Gen.Physiol., 153, 2021
|
|
5TSE
 
 | | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii. | | Descriptor: | FORMIC ACID, LPS-assembly lipoprotein LptE | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii.
To Be Published
|
|
5U0D
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-(2,4-dimethoxyphenyl)-2-sulfanylidene-1,3-oxazolidin-4-one, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
7KZZ
 
 | | Cryo-EM structure of YiiP-Fab complex in Holo state | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2R heavy chain, Fab2R light chain, ... | | Authors: | Lopez-Redondo, M.L, Fan, S, Koide, A, Koide, S, Beckstein, O, Stokes, D.L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-06 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP.
J.Gen.Physiol., 153, 2021
|
|
3KBC
 
 | | Crystal structure of GltPh K55C-A364C mutant crosslinked with divalent mercury | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, MERCURY (II) ION, ... | | Authors: | Reyes, N, Ginter, C, Boudker, O. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Transport mechanism of a bacterial homologue of glutamate transporters.
Nature, 462, 2009
|
|
5U8T
 
 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | Descriptor: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | Authors: | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TR7
 
 | | Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Shatsman, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative D-alanyl-D-alanine carboxypeptidase from Vibrio cholerae O1 biovar eltor str. N16961
To Be Published
|
|
5UBD
 
 | | Crystal structure of the N-terminal domain (domain 1) of RctB, RctB-1-124-L48M | | Descriptor: | RctB replication initiator protein | | Authors: | Orlova, N, Ivashkiv, O, Waldor, M.K, Jeruzalmi, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The replication initiator of the cholera pathogen's second chromosome shows structural similarity to plasmid initiators.
Nucleic Acids Res., 45, 2017
|
|
5TY8
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-phenyl-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
1BVT
 
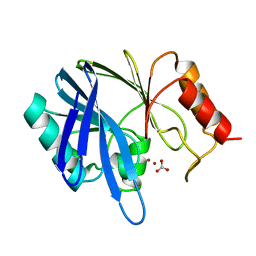 | | METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS 569/H/9 | | Descriptor: | BICARBONATE ION, PROTEIN (BETA-LACTAMASE), ZINC ION | | Authors: | Carfi, A, Duee, E, Dideberg, O. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 A resolution structure of the zinc (II) beta-lactamase from Bacillus cereus.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3KBO
 
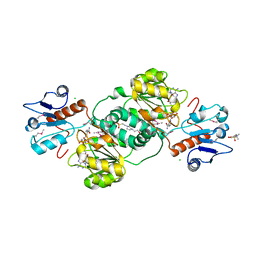 | | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Glyoxylate/hydroxypyruvate reductase A, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP.
To be Published
|
|
3KZF
 
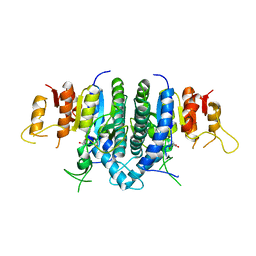 | | Structure of Giardia Carbamate Kinase | | Descriptor: | Carbamate kinase, GLYCEROL | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-12-08 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure and characterization of carbamate kinase from the human parasite Giardia lamblia.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5TP0
 
 | | Human mesotrypsin in complex with diminazene | | Descriptor: | BERENIL, CALCIUM ION, SULFATE ION, ... | | Authors: | Kayode, O, Soares, A, Radisky, E.S. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule inhibitors of mesotrypsin from a structure-based docking screen.
PLoS ONE, 12, 2017
|
|
5TRO
 
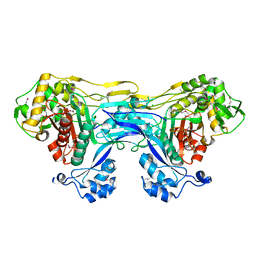 | | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 1 | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus.
To Be Published
|
|
3KIH
 
 | | The crystal structures of two fragments truncated from 5-bladed beta-propeller lectin, tachylectin-2 (Lib2-D2-15) | | Descriptor: | 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 5-bladed beta-propeller lectin | | Authors: | Dym, O, Tawfik, D.S, Yadid, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-11-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metamorphic proteins mediate evolutionary transitions of structure
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1CPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN (PH 10), SODIUM ION | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
5U4H
 
 | | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, FORMIC ACID, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid.
To Be Published
|
|
3KHH
 
 | | Dpo4 extension ternary complex with a C base opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-AMINOFLUORENE, 5'-D(*CP*CP*TP*A*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KIF
 
 | | The crystal structures of two fragments truncated from 5-bladed beta-propeller lectin, tachylectin-2 (Lib1-B7-18 and Lib2-D2-15) | | Descriptor: | 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 5-bladed beta-propeller lectin, SULFATE ION | | Authors: | Dym, O, Tawfik, D.S, Yadid, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-11-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metamorphic proteins mediate evolutionary transitions of structure
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KKU
 
 | | Cruzain in complex with a non-covalent ligand | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, N-[2-(1H-benzimidazol-2-yl)ethyl]-2-(2-bromophenoxy)acetamide, ... | | Authors: | Ferreira, R.S, Eidam, O, Shoichet, B.K. | | Deposit date: | 2009-11-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors.
J.Med.Chem., 53, 2010
|
|
