8CBJ
 
 | | Cryo-EM structure of Otu2-bound cytoplasmic pre-40S ribosome biogenesis complex | | Descriptor: | 20S pre-ribosomal RNA, 20S-pre-rRNA D-site endonuclease NOB1, 40S ribosomal protein S0-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CAS
 
 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
1QPY
 
 | | CRYSTAL STRUCTURE OF BACKBONE MODIFIED PNA HEXAMER | | Descriptor: | PEPTIDE NUCLEIC ACID 5'-(*CP1*GPN*TP1*APN*CP1*GPN*LYS)-3' | | Authors: | Haima, G, Rasmussen, H, Schmidt, G, Jensen, D.K, Kastrup, J.S, Stafshede, P.W, Norden, B, Buchardt, O, Nielsen, P.E. | | Deposit date: | 1999-05-14 | | Release date: | 2001-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide Nucleic Acids (PNA) derived from N-(N-methylaminoethyl)glycine. Synthesis, hybridization and structural properties
New J.Chem., 23, 1999
|
|
8IGI
 
 | | Crystal structure of HP1526 (XthA)- a base excision DNA repair protein in Helicobacter pylori | | Descriptor: | 1,3-BUTANEDIOL, Exodeoxyribonuclease (LexA), MANGANESE (II) ION | | Authors: | Dinh, T.T, Dao, O, Lee, K.H. | | Deposit date: | 2023-02-20 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease XthA (HP1526 protein) from Helicobacter pylori.
Biochem.Biophys.Res.Commun., 663, 2023
|
|
1QXN
 
 | | Solution Structure of the 30 kDa Polysulfide-sulfur Transferase Homodimer from Wolinella Succinogenes | | Descriptor: | PENTASULFIDE-SULFUR, sulfide dehydrogenase | | Authors: | Lin, Y.J, Dancea, F, Loehr, F, Klimmek, O, Pfeiffer-Marek, S, Nilges, M, Wienk, H, Kroeger, A, Rueterjans, H. | | Deposit date: | 2003-09-08 | | Release date: | 2004-02-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 30 kDa Polysulfide-Sulfur Transferase Homodimer from Wolinella succinogenes
Biochemistry, 43, 2004
|
|
1QX6
 
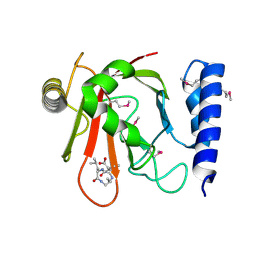 | | Crystal structure of Sortase B complexed with E-64 | | Descriptor: | N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, NPQTN specific sortase B | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-04 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
1QXA
 
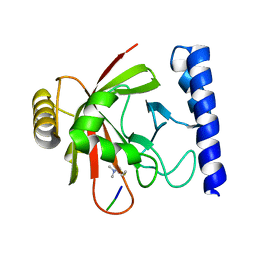 | | Crystal structure of Sortase B complexed with Gly3 | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, NPQTN specific sortase B, peptide GLY-GLY-GLY | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-05 | | Release date: | 2004-04-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
1QXD
 
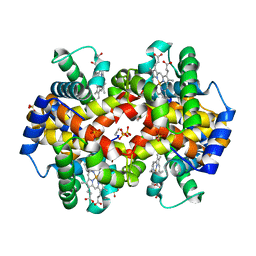 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
1QGJ
 
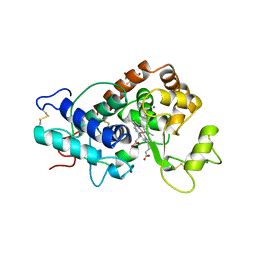 | | ARABIDOPSIS THALIANA PEROXIDASE N | | Descriptor: | CALCIUM ION, GLUTATHIONE, PEROXIDASE N, ... | | Authors: | Mirza, O, Oestergaard, L, Welinder, K.G, Henriksen, A, Gajhede, M. | | Deposit date: | 1999-04-29 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arabidopsis thaliana peroxidase N: structure of a novel neutral peroxidase.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6TEO
 
 | | Crystal structure of a yeast Snu114-Prp8 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor 8, ... | | Authors: | Ganichkin, O, Jia, J, Loll, B, Absmeier, E, Wahl, M.C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Snu114-GTP-Prp8 module forms a relay station for efficient splicing in yeast.
Nucleic Acids Res., 48, 2020
|
|
1QDB
 
 | | CYTOCHROME C NITRITE REDUCTASE | | Descriptor: | CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, HEME C, ... | | Authors: | Einsle, O, Messerschmidt, A, Stach, P, Huber, R, Kroneck, P.M.H. | | Deposit date: | 1999-05-19 | | Release date: | 1999-08-18 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of cytochrome c nitrite reductase.
Nature, 400, 1999
|
|
8J1J
 
 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Minimal and most efficient genome editing Cas enzyme
To Be Published
|
|
8J3R
 
 | | Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (37-MER), DNA (38-MER), MAGNESIUM ION, ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Minimal and most efficient genome editing Cas enzyme
To Be Published
|
|
1P93
 
 | | CRYSTAL STRUCTURE OF THE AGONIST FORM OF GLUCOCORTICOID RECEPTOR | | Descriptor: | DEXAMETHASONE, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2003-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
8JEK
 
 | | Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
8CUD
 
 | |
8JEJ
 
 | | Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
8CUJ
 
 | |
8CV3
 
 | |
8CTD
 
 | |
8CV2
 
 | |
8CUI
 
 | |
8CTC
 
 | |
8CUA
 
 | |
8J97
 
 | | Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local refine, cross-linked) | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sano, F.K, Shihoya, W, Banerjee, R, Nureki, O, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
