7CON
 
 | |
7CP8
 
 | |
1P83
 
 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | Descriptor: | 10 kDa chaperonin | | Authors: | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | Deposit date: | 2003-05-06 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6O4X
 
 | | Binary complex of native hAChE with 9-aminoacridine | | Descriptor: | 9-AMINOACRIDINE, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
6O5S
 
 | |
2M3Z
 
 | |
7E7Z
 
 | |
7B16
 
 | | TRPC4 in complex with inhibitor GFB-9289 | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 5-chloranyl-4-(4-cyclohexyl-3-oxidanylidene-piperazin-1-yl)-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7E83
 
 | |
7E87
 
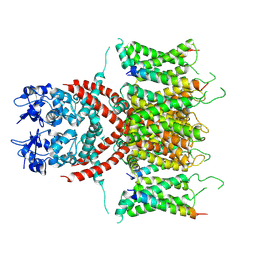 | |
7E84
 
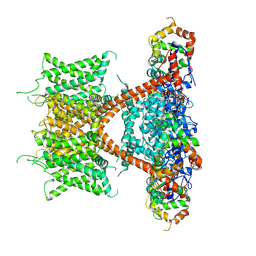 | | CryoEM structure of human Kv4.2-KChIP1 complex | | Descriptor: | Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-02-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8E
 
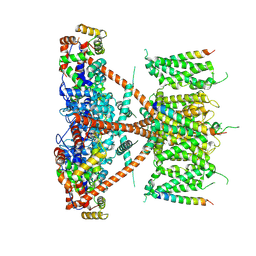 | |
7B0S
 
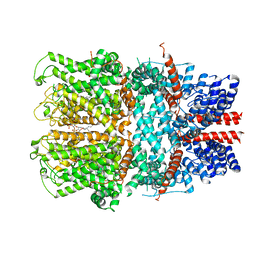 | | TRPC4 in complex with inhibitor GFB-8438 | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 5-chloranyl-4-[3-oxidanylidene-4-[[2-(trifluoromethyl)phenyl]methyl]piperazin-1-yl]-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7E8G
 
 | |
7E89
 
 | |
7E8B
 
 | | CryoEM structure of human Kv4.2-DPP6S complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8H
 
 | | CryoEM structure of human Kv4.2-DPP6S-KChIP1 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8D
 
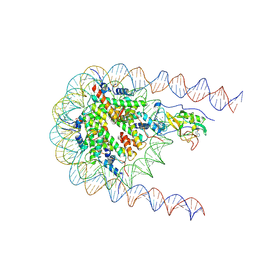 | | NSD2 E1099K mutant bound to nucleosome | | Descriptor: | DNA (185-MER), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Sengoku, T, Sato, K, Nishizawa, T, Nureki, O, Ogata, K. | | Deposit date: | 2021-03-01 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the regulation of the normal and oncogenic methylation of nucleosomal histone H3 Lys36 by NSD2.
Nat Commun, 12, 2021
|
|
7B1G
 
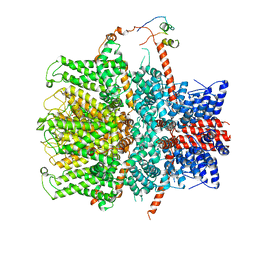 | | TRPC4 in complex with Calmodulin | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, CALCIUM ION, Calmodulin-1, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B0J
 
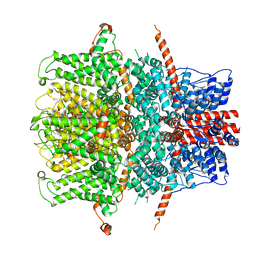 | | TRPC4 in LMNG detergent | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, Transient receptor potential cation channel subfamily c member 4a | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B05
 
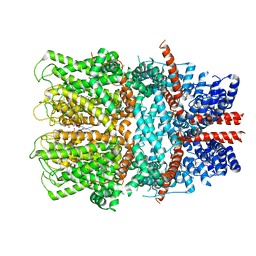 | | TRPC4 in complex with inhibitor GFB-8749 | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4-[4-[[4,4-bis(fluoranyl)cyclohexyl]methyl]-3-oxidanylidene-piperazin-1-yl]-5-chloranyl-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7D7Q
 
 | | Crystal structure of the transmembrane domain and linker region of Salpingoeca rosetta rhodopsin phosphodiesterase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphodiesterase, RETINAL | | Authors: | Ikuta, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into the mechanism of rhodopsin phosphodiesterase.
Nat Commun, 11, 2020
|
|
7E46
 
 | |
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
