4UWO
 
 | | Native di-zinc VIM-26. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | GLYCEROL, METALLO-BETA-LACTAMASE VIM-26, SODIUM ION, ... | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
1ZDV
 
 | | Solution Structure of the type 1 pilus assembly platform FimD(25-139) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
4UWR
 
 | | Mono-zinc VIM-26. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | FORMIC ACID, METALLO-BETA-LACTAMASE VIM-26, SODIUM ION, ... | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
4V1H
 
 | |
4WIB
 
 | | Crystal structure of Magnesium transporter MgtE | | Descriptor: | CALCIUM ION, Magnesium transporter MgtE | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-09-25 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
4WH5
 
 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, lincomycin-bound | | Descriptor: | CHLORIDE ION, LINCOMYCIN, Lincosamide resistance protein, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, O, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-20 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | CRYSTAL STRUCTURE OF LINCOSAMIDE ANTIBIOTIC ADENYLYLTRANSFERASE LNUA, LINCOMYCIN BOUND
To Be Published
|
|
4WOE
 
 | | The duplicated taurocyamine kinase from Schistosoma mansoni with bound transition state analog (TSA) components | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
4WN4
 
 | | Crystal structure of designed cPPR-polyA protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4WOT
 
 | | ROCK2 IN COMPLEX WITH 1426382-07-1 | | Descriptor: | Rho-associated protein kinase 2, methyl 3-[({2'-(aminomethyl)-5'-[(3-fluoropyridin-4-yl)carbamoyl]biphenyl-3-yl}carbonyl)amino]-4-fluorobenzoate | | Authors: | Augustin, M, Krapp, S, Boland, S, Defert, O, Bourin, A, Alen, J, Leysen, D. | | Deposit date: | 2014-10-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Design, synthesis, and biological evaluation of novel, highly active soft ROCK inhibitors.
J. Med. Chem., 58, 2015
|
|
4V6M
 
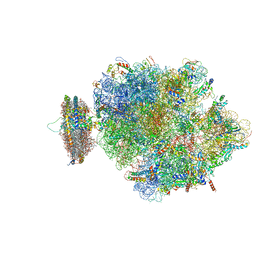 | | Structure of the ribosome-SecYE complex in the membrane environment | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | Authors: | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | Deposit date: | 2011-02-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4V6X
 
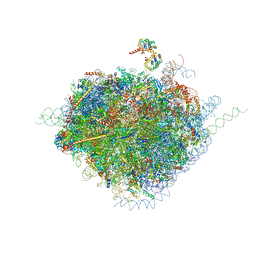 | | Structure of the human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
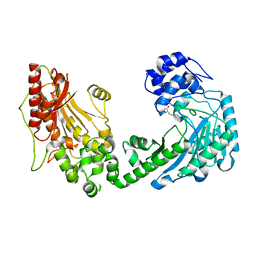
4X3B
 
 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
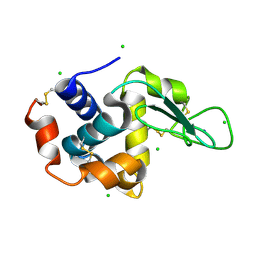
4WOD
 
 | | The duplicated taurocyamine kinase from Schistosoma mansoni complexed with arginine | | Descriptor: | ARGININE, Taurocyamine kinase | | Authors: | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
4X2S
 
 | |
4WKZ
 
 | | Complex of autonomous ScaG cohesin CohG and X-doc domains | | Descriptor: | ACETATE ION, Autonomous cohesin, CALCIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Bayer, E.A, Yaniv, O, Shimon, L.J.W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Complex of autonomous ScaG cohesin CohG and X-doc domains
To Be Published
|
|
4X5R
 
 | | Crystal structure of FimH in complex with a squaryl-phenyl alpha-D-mannopyranoside derivative | | Descriptor: | 2-chloro-4-{[2-(4-methylpiperazin-1-yl)-3,4-dioxocyclobut-1-en-1-yl]amino}phenyl alpha-D-mannopyranoside, Protein FimH, SULFATE ION | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4WNA
 
 | | Structure of the Nitrogenase MoFe Protein from Azotobacter vinelandii Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Zhang, L, Einsle, O, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
4X5P
 
 | | Crystal structure of FimH in complex with a benzoyl-amidophenyl alpha-D-mannopyranoside | | Descriptor: | 4-{[3-chloro-4-(alpha-D-mannopyranosyloxy)phenyl]carbamoyl}benzoic acid, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.997 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4WRP
 
 | | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Cuff, M.E, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To Be Published
|
|
4WSL
 
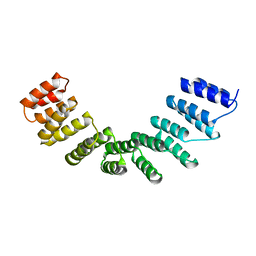 | | Crystal structure of designed cPPR-polyC protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
1QME
 
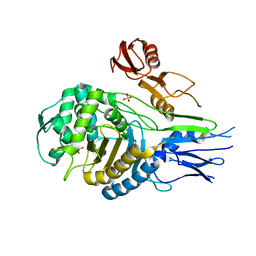 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1OVD
 
 | | THE K136E MUTANT OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE A IN COMPLEX WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-03-26 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function.
J.Biol.Chem., 278, 2003
|
|
1S46
 
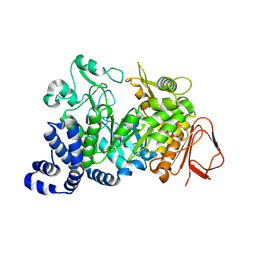 | | Covalent intermediate of the E328Q amylosucrase mutant | | Descriptor: | amylosucrase, beta-D-glucopyranose | | Authors: | Jensen, M.H, Mirza, O, Albenne, C, Remaud-Simeon, M, Monsan, P, Gajhede, M, Skov, L.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the covalent intermediate of amylosucrase from Neisseria polysaccharea.
Biochemistry, 43, 2004
|
|
1PRC
 
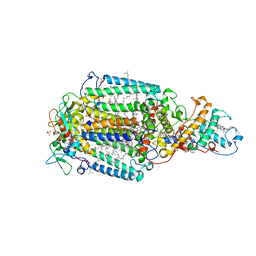 | | CRYSTALLOGRAPHIC REFINEMENT AT 2.3 ANGSTROMS RESOLUTION AND REFINED MODEL OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS | | Descriptor: | 15-trans-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Deisenhofer, J, Epp, O, Miki, K, Huber, R, Michel, H. | | Deposit date: | 1988-02-04 | | Release date: | 1989-01-09 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic refinement at 2.3 A resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis.
J.Mol.Biol., 246, 1995
|
|
1ZS2
 
 | | Amylosucrase Mutant E328Q in a ternary complex with sucrose and maltoheptaose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, van der Veen, B.A, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the Glu328Gln mutant of Neisseria polysaccharea amylosucrase in complex with sucrose and maltoheptaose
BIOCATAL.BIOTRANSFOR., 24, 2006
|
|
