4F13
 
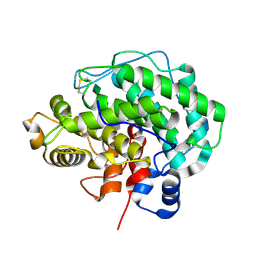 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6XWW
 
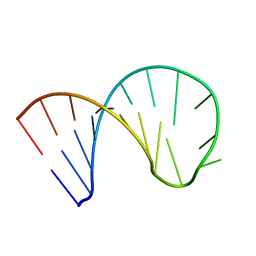 | |
6XX1
 
 | | The unique CBM3-Clocl_1192 of Hungateiclostridium clariflavum | | Descriptor: | Cellulose binding domain-containing protein | | Authors: | Milana, M.V, Almog, R, Yaniv, O, Oded, L, Inna, R.G, Felix, F, Edward, A.B, Raphael, L. | | Deposit date: | 2020-01-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The unique CBM3-Cthe_0271 from Ruminoclostridium thermocellum and CBM3-Clocl_1192 from Hungateiclostridium clariflavum
To Be Published
|
|
5MQ9
 
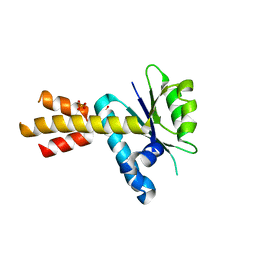 | | Crystal structure of Rae1 (YacP) from Bacillus subtilis (W164L mutant) | | Descriptor: | SULFATE ION, Uncharacterized protein YacP | | Authors: | Piton, J, Gilet, L, Pellegrini, O, Leroy, M, Figaro, S, Condon, C. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.174 Å) | | Cite: | Rae1/YacP, a new endoribonuclease involved in ribosome-dependent mRNA decay in Bacillus subtilis.
EMBO J., 36, 2017
|
|
1CS2
 
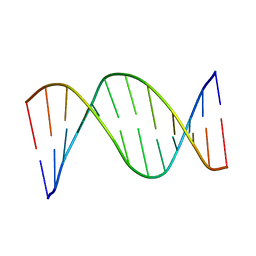 | | NMR STRUCTURES OF B-DNA D(CTACTGCTTTAG).D(CTAAAGCAGTAG) | | Descriptor: | 5'-d(*CP*TP*AP*AP*AP*GP*CP*AP*GP*TP*AP*G)-3', 5'-d(*CP*TP*AP*CP*TP*GP*CP*TP*TP*TP*AP*G)-3' | | Authors: | Leporc, S, Mauffret, O, Tevanian, G, Lescot, E, Monnot, M, Fermandjian, S. | | Deposit date: | 1999-08-16 | | Release date: | 1999-08-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR and molecular modelling analysis of d(CTACTGCTTTAG). d(CTAAAGCAGTAG) reveals that the particular behaviour of TpA steps is related to edge-to-edge contacts of their base-pairs in the major groove
Nucleic Acids Res., 27, 1999
|
|
1CEE
 
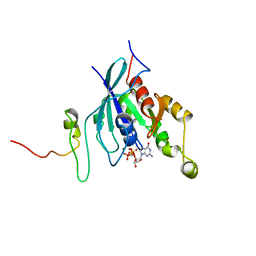 | | SOLUTION STRUCTURE OF CDC42 IN COMPLEX WITH THE GTPASE BINDING DOMAIN OF WASP | | Descriptor: | GTP-BINDING RHO-LIKE PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Abdul-Manan, N, Aghazadeh, B, Liu, G.A, Majumdar, A, Ouerfelli, O, Rosen, M.K. | | Deposit date: | 1999-03-08 | | Release date: | 1999-06-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cdc42 in complex with the GTPase-binding domain of the 'Wiskott-Aldrich syndrome' protein.
Nature, 399, 1999
|
|
6XTJ
 
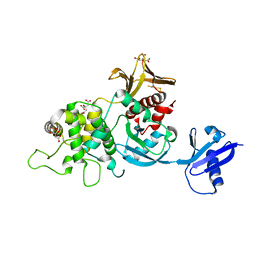 | | The high resolution structure of the FERM domain of human FERMT2 | | Descriptor: | CITRIC ACID, Fermitin family homolog 2,Fermitin family homolog 2,Fermitin family homolog 2 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structure of the FERM domain of human FERMT2
To Be Published
|
|
5MVJ
 
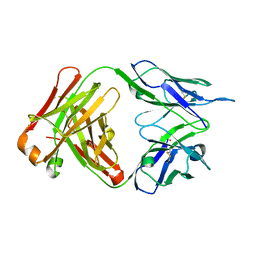 | |
6XY7
 
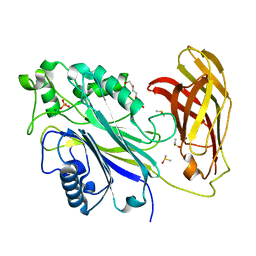 | | Human SHIP1 with magnesium and phosphate bound to the active site | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bradshaw, W.J, Scacioc, A, Fernandez-Cid, A, Mckinley, G, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-29 | | Release date: | 2020-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5MKP
 
 | | Non redox thiolation in transfer RNAs occuring via sulfur activation by a [4Fe-4S] cluster | | Descriptor: | Fe4 H S5, PH0300, ZINC ION | | Authors: | Arragain, S, Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2016-12-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonredox thiolation in tRNA occurring via sulfur activation by a [4Fe-4S] cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5MM4
 
 | |
4F4I
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form | | Descriptor: | Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form
To be Published
|
|
1DIK
 
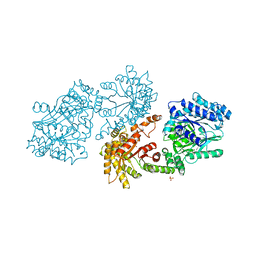 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C.H. | | Deposit date: | 1995-12-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4F86
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with GPP and sinefungin | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5MR1
 
 | | Crystal structure of the Pleckstrin homology domain of Interactor protein for cytohesin exchange factors 1 (IPCEF1) | | Descriptor: | Interactor protein for cytohesin exchange factors 1 | | Authors: | Newman, J.A, Aitkenhead, H, Wang, D, Burgess-Brown, N, Williams, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the Pleckstrin homology domain of Interactor protein for cytohesin exchange factors 1 (IPCEF1)
To be published
|
|
1DBU
 
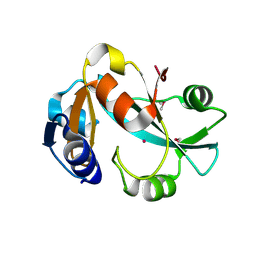 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase protein from H. influenzae (HI1434) | | Descriptor: | MERCURY (II) ION, cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
5MTV
 
 | | Active structure of EHD4 complexed with ATP-gamma-S | | Descriptor: | EH domain-containing protein 4, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Melo, A.A, Daumke, O. | | Deposit date: | 2017-01-10 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into the activation mechanism of dynamin-like EHD ATPases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4F85
 
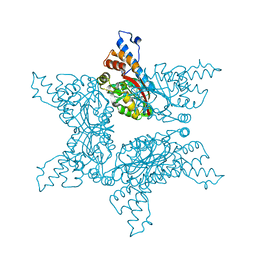 | | Structure analysis of Geranyl diphosphate methyltransferase | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1DPK
 
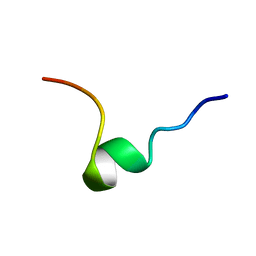 | |
4GCW
 
 | | Crystal structure of RNase Z in complex with precursor tRNA(Thr) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ribonuclease Z, TRNA(THR), ... | | Authors: | Pellegrini, O, Li de la Sierra-Gallay, I, Piton, J, Gilet, L, Condon, C. | | Deposit date: | 2012-07-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation of tRNA Maturation by Downstream Uracil Residues in B. subtilis
Structure, 20, 2012
|
|
4G5J
 
 | | Crystal structure of EGFR kinase in complex with BIBW2992 | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)but-2-enamide, Epidermal growth factor receptor, N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)butanamide | | Authors: | Solca, F, Dahl, G, Zoephel, A, Bader, G, Sanderson, M, Klein, C, Kraemer, O, Himmelsbach, F, Haaksma, E, Adolf, G.R. | | Deposit date: | 2012-07-18 | | Release date: | 2012-08-29 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Target Binding Properties and Cellular Activity of Afatinib (BIBW 2992), an Irreversible ErbB Family Blocker.
J.Pharmacol.Exp.Ther., 343, 2012
|
|
4G65
 
 | | Potassium transporter peripheral membrane component (trkA) from Vibrio vulnificus | | Descriptor: | CHLORIDE ION, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-18 | | Release date: | 2012-08-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Potassium transporter peripheral membrane component (trkA) from Vibrio vulnificus.
To be Published
|
|
5MAQ
 
 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ADP and PPi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4GD5
 
 | | X-ray Crystal Structure of a Putative Phosphate ABC Transporter Substrate-Binding Protein with Bound Phosphate from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray Crystal Structure of a Putative Phosphate ABC Transporter Substrate-Binding Protein with Bound Phosphate from Clostridium perfringens
To be Published
|
|
