7OI0
 
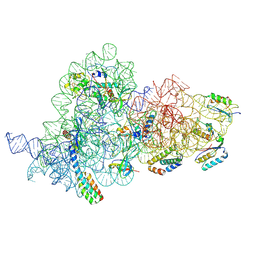 | | E.coli delta rbfA pre-30S ribosomal subunit class D | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Maksimova, E, Korepanov, A, Baymukhametov, T, Kravchenko, O, Stolboushkina, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center.
Int J Mol Sci, 22, 2021
|
|
2BIJ
 
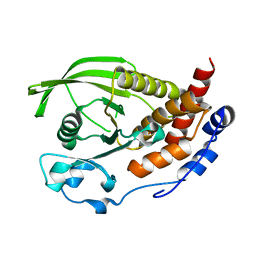 | | Crystal structure of the human protein tyrosine phosphatase PTPN5 (STEP, striatum enriched enriched Phosphatase) | | Descriptor: | SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE, NON-RECEPTOR TYPE 5 | | Authors: | Barr, A.J, Debreczeni, J.E, Eswaran, J, Smee, C, Burgess, N, Gileadi, O, Sundstrom, M, Arrowsmith, C, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases.
Biochem. J., 395, 2006
|
|
2CLP
 
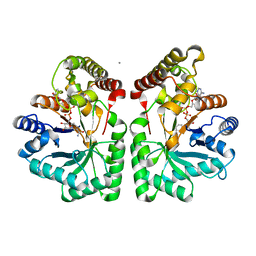 | | Crystal structure of human aflatoxin B1 aldehyde reductase member 3 | | Descriptor: | AFLATOXIN B1 ALDEHYDE REDUCTASE MEMBER 3, CALCIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Debreczeni, J.E, Marsden, B.D, Johansson, C, Kavanagh, K, Guo, K, Smee, C, Gileadi, O, Turnbull, A, Papagrigoriou, E, von Delft, F, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Oppermann, U. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Aflatoxin B1 Aldehyde Reductase Member 3
To be Published
|
|
2C7S
 
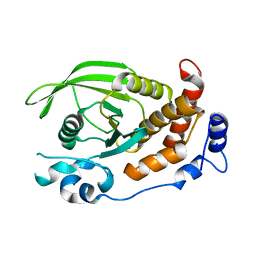 | | Crystal structure of human protein tyrosine phosphatase kappa at 1.95A resolution | | Descriptor: | ACETATE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE KAPPA | | Authors: | Debreczeni, J.E, Ugochukwu, E, Eswaran, J, Barr, A, Das, S, Burgess, N, Gileadi, O, Longman, E, von Delft, F, Knapp, S, Sundstron, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-11-28 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
2CNJ
 
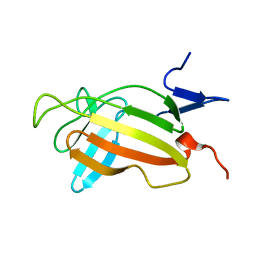 | | NMR studies on the interaction of Insulin-Growth Factor II (IGF-II) with IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Williams, C, Prince, S, Zaccheo, O, Hassan, A.B, Crosby, J, Crump, M. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights Into the Interaction of Insulin-Like Growth Factor 2 with Igf2R Domain 11.
Structure, 15, 2007
|
|
7OTL
 
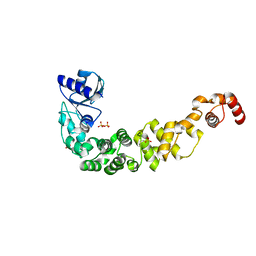 | | Structure of a psychrophilic CCA-adding enzyme crystallized by counter-diffusion | | Descriptor: | CCA-adding enzyme, SULFATE ION | | Authors: | Rollet, K, de Wijn, R, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2021-06-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
7OQX
 
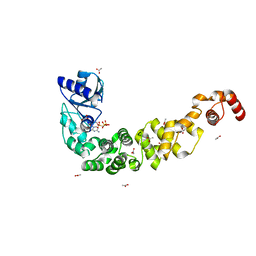 | | Crystal structure of a psychrophilic CCA-adding enzyme in complex with CMPcPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ACETATE ION, CCA-adding enzyme, ... | | Authors: | Rollet, K, de Wijn, R, Bluhm, A, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
7OTR
 
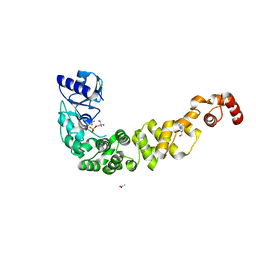 | | Crystal structure of a psychrophilic CCA-adding enzyme determined by SAD phasing | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CCA-adding protein, ... | | Authors: | Rollet, K, de Wijn, R, Rios-Santacruz, R, Hennig, O, Betat, H, Moerl, M, Sauter, C. | | Deposit date: | 2021-06-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
2BWJ
 
 | | Structure of adenylate kinase 5 | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE 5, CHLORIDE ION, ... | | Authors: | Bunkoczi, G, Filippakopoulos, P, Fedorov, O, Jansson, A, Longman, E, Ugochukwu, E, Knapp, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Adenylate Kinase 5
To be Published
|
|
2BV5
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PROTEIN TYROSINE PHOSPHATASE PTPN5 AT 1.8A RESOLUTION | | Descriptor: | GLYCEROL, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE, ... | | Authors: | Debreczeni, J.E, Barr, A.J, Eswaran, J, Smee, C, Burgess, N, Gileadi, O, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-06-22 | | Release date: | 2005-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases.
Biochem. J., 395, 2006
|
|
2BR9
 
 | | 14-3-3 Protein Epsilon (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN EPSILON, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Yang, X, Elkins, J.M, Soundararajan, M, Fedorov, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A. | | Deposit date: | 2005-05-03 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2BQ0
 
 | | 14-3-3 Protein Beta (Human) | | Descriptor: | 14-3-3 BETA/ALPHA | | Authors: | Yang, X, Elkins, J.M, Fedorov, O, Longman, E.J, Sobott, L, Ball, L.J, Sundstrom, M, Arrowsmith, C, Edwards, A, Doyle, D.A. | | Deposit date: | 2005-04-26 | | Release date: | 2005-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2B46
 
 | | Crystal structure of an engineered uninhibited Bacillus subtilis xylanase in substrate bound state | | Descriptor: | Endo-1,4-beta-xylanase A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Sorensen, J.F, Sibbesen, O, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase
To be Published
|
|
2B45
 
 | | Crystal structure of an engineered uninhibited Bacillus subtilis xylanase in free state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Sorensen, J.F, Sibbesen, O, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase
To be Published
|
|
2A3K
 
 | | Crystal Structure of the Human Protein Tyrosine Phosphatase, PTPN7 (HePTP, Hematopoietic Protein Tyrosine Phosphatase) | | Descriptor: | PHOSPHATE ION, protein tyrosine phosphatase, non-receptor type 7, ... | | Authors: | Barr, A, Turnbull, A.P, Das, S, Eswaran, J, Debreczeni, J.E, Longmann, E, Smee, C, Burgess, N, Gileadi, O, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-24 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
2AHS
 
 | | Crystal Structure of the Catalytic Domain of Human Tyrosine Receptor Phosphatase Beta | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta, ... | | Authors: | Ugochukwu, E, Eswaran, J, Barr, A, Gileadi, O, Sobott, F, Burgess, N, Ball, L, Bray, J, von Delft, F, Debreczeni, J, Bunkoczi, G, Turnbull, A, Das, S, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2CJZ
 
 | | crystal structure of the c472s mutant of human protein tyrosine phosphatase ptpn5 (step, striatum enriched phosphatase) in complex with phosphotyrosine | | Descriptor: | 1,2-ETHANEDIOL, HUMAN PROTEIN TYROSINE PHOSPHATASE PTPN5, O-PHOSPHOTYROSINE | | Authors: | Debreczeni, J.E, Barr, A.J, Eswaran, J, Smee, C, Burgess, N, Gileadi, O, Savitsky, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, Knapp, S, von Delft, F. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2QXW
 
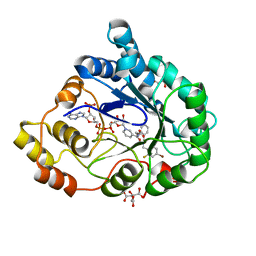 | | Perdeuterated alr2 in complex with idd594 | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O, Cousido-Siah, A, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1DYM
 
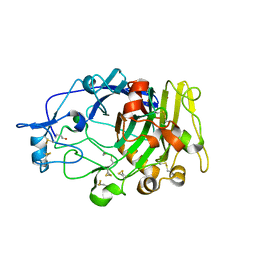 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
5HL3
 
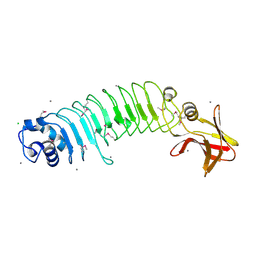 | | Crystal structure of Listeria monocytogenes InlP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lmo2470 protein | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Kiryukhina, O, Kwon, K, Faralla, C, Bakardjiev, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing.
Plos Pathog., 14, 2018
|
|
1C58
 
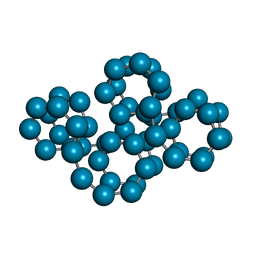 | | CRYSTAL STRUCTURE OF CYCLOAMYLOSE 26 | | Descriptor: | Cyclohexacosakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Gessler, K, Saenger, W, Nimz, O. | | Deposit date: | 1999-11-04 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | V-Amylose at atomic resolution: X-ray structure of a cycloamylose with 26 glucose residues (cyclomaltohexaicosaose).
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
2VN9
 
 | | Crystal Structure of Human Calcium Calmodulin dependent Protein Kinase II delta isoform 1, CAMKD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II DELTA CHAIN, CHLORIDE ION, ... | | Authors: | Roos, A.K, Rellos, P, Salah, E, Pike, A.C.W, Fedorov, O, Pilka, E.S, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Camkiidelta/Calmodulin Complex Reveals the Molecular Mechanism of Camkii Kinase Activation.
Plos Biol., 8, 2010
|
|
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
2UZC
 
 | | Structure of human PDLIM5 in complex with the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ AND LIM DOMAIN 5 | | Authors: | Bunkoczi, G, Elkins, J, Salah, E, Burgess-Brown, N, Papagrigoriou, E, Pike, A.C.W, Turnbull, A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D. | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms.
Protein Sci., 19, 2010
|
|
