7L6Y
 
 | |
1DJX
 
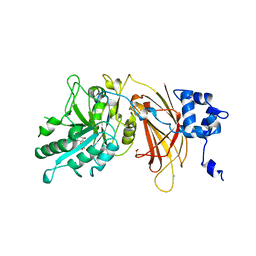 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-1,4,5-TRISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
7L6Z
 
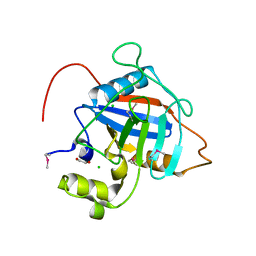 | | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6
To Be Published
|
|
7L71
 
 | |
1EBA
 
 | |
3L6C
 
 | | X-ray crystal structure of rat serine racemase in complex with malonate a potent inhibitor | | Descriptor: | MALONATE ION, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, M.A, Mack, V, Ebneth, A, Moraes, I, Felicetti, B, Wood, M, Schonfeld, D, Mather, O, Cesura, A, Barker, J. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
1E8X
 
 | | STRUCTURAL INSIGHTS INTO PHOSHOINOSITIDE 3-KINASE ENZYMATIC MECHANISM AND SIGNALLING | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LUTETIUM (III) ION, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Perisic, O, Ried, C, Stephens, L, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
3LT3
 
 | | Crystal structure of Rv3671c from M. tuberculosis H37Rv, Ser343Ala mutant, inactive form | | Descriptor: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | Authors: | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | Deposit date: | 2010-02-14 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
5UKM
 
 | | bovine GRK2 in complex with human Gbetagamma subunits and CCG258208 (14as) | | Descriptor: | 5-[(3S,4R)-3-{[(2H-1,3-benzodioxol-5-yl)oxy]methyl}piperidin-4-yl]-2-fluoro-N-[(1H-pyrazol-5-yl)methyl]benzamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cruz-Rodriguez, O, Tesmer, J.J.G. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure-Based Design of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors Based on Paroxetine.
J. Med. Chem., 60, 2017
|
|
3L5L
 
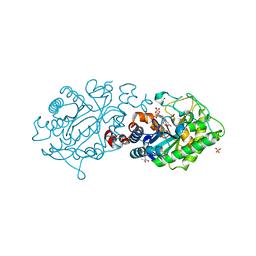 | | Xenobiotic Reductase A - oxidized | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-22 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
3L68
 
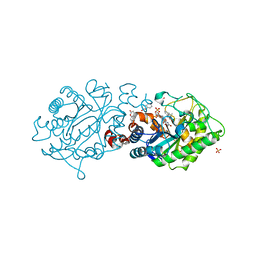 | | Xenobiotic Reductase A - C25S variant with coumarin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
5UY6
 
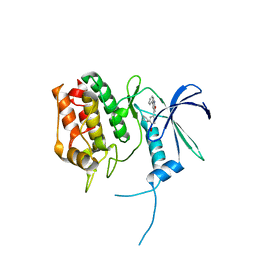 | | Crystal Structure of the Human CAMKK2B | | Descriptor: | 2-cyclopentyl-4-(5-phenylfuro[2,3-b]pyridin-3-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Counago, R.M, Dewry, D, Bountra, C, Arruda, P, Edwards, A.M, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Human CAMKK2B
To Be Published
|
|
7LYZ
 
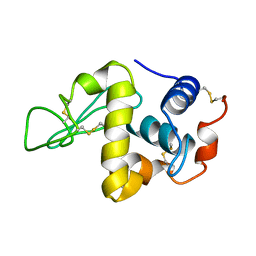 | | PROTEIN MODEL BUILDING BY THE USE OF A CONSTRAINED-RESTRAINED LEAST-SQUARES PROCEDURE | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Moult, J, Yonath, A, Sussman, J, Herzberg, O, Podjarny, A, Traub, W. | | Deposit date: | 1977-05-06 | | Release date: | 1977-06-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Model Building by the Use of a Constrained-Restrained Least-Squares Procedure
J.Appl.Crystallogr., 16, 1983
|
|
3LGR
 
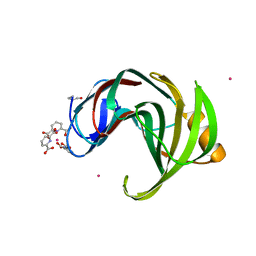 | | Xylanase II from Trichoderma reesei cocrystallized with tris-dipicolinate europium | | Descriptor: | EUROPIUM ION, Endo-1,4-beta-xylanase 2, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Pompidor, G, Kahn, R, Maury, O. | | Deposit date: | 2010-01-21 | | Release date: | 2011-01-19 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5UKF
 
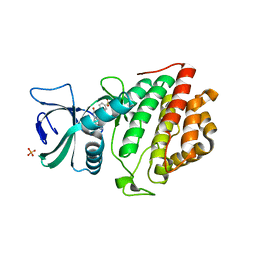 | | Crystal Structure of the Human Vaccinia-related Kinase 1 Bound to an Oxindole Inhibitor | | Descriptor: | 4-{[(Z)-(7-oxo-6,7-dihydro-8H-[1,3]thiazolo[5,4-e]indol-8-ylidene)methyl]amino}benzene-1-sulfonamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, Wells, C, Zuercher, W, Willson, T.M, Bountra, C, Edwards, A.M, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-22 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
3L9I
 
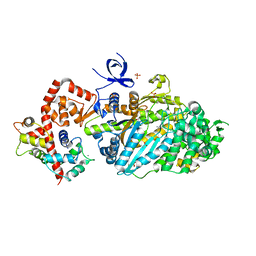 | | Myosin VI nucleotide-free (mdinsert2) L310G mutant crystal structure | | Descriptor: | ACETATE ION, CALCIUM ION, Calmodulin, ... | | Authors: | Pylypenko, O, Song, L, Sweeney, L.H, Houdusse, A. | | Deposit date: | 2010-01-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of insert i of myosin VI in modulating nucleotide affinity
To be Published
|
|
1DJY
 
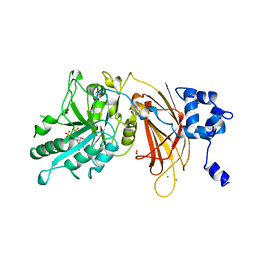 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2,4,5-TRISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-2,4,5-TRIPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
3L66
 
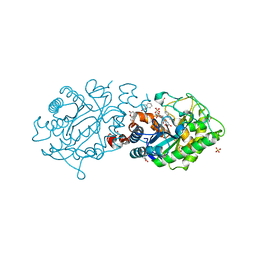 | | Xenobiotic Reductase A - C25A Variant with Coumarin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
3L5M
 
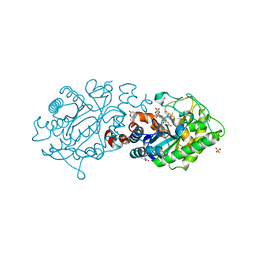 | | Xenobiotic reductase A - coumarin bound | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-22 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
3LLB
 
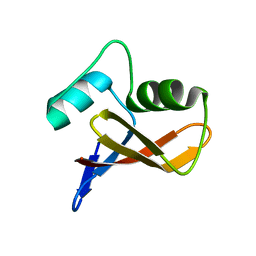 | | The crystal structure of the protein PA3983 with unknown function from Pseudomonas aeruginosa PAO1 | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, R, Kagan, O, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the protein NE1376 with unknown function from Nitrosomonas europaea ATCC 19718
To be Published
|
|
3L67
 
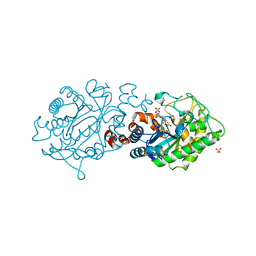 | | Xenobiotic reductase A - C25S variant | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
3LHH
 
 | | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain protein | | Authors: | Tan, K, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1.
To be Published
|
|
3L65
 
 | | Xenobiotic Reductase A - C25A Mutant | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
5VZW
 
 | | TRIM23 RING domain in complex with UbcH5-Ub | | Descriptor: | E3 ubiquitin-protein ligase TRIM23, Ubiquitin, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Pornillos, O, Dawidziak, D. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase.
Proteins, 85, 2017
|
|
3LPW
 
 | | Crystal structure of the FnIII-tandem A77-A78 from the A-band of titin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, A77-A78 domain from Titin | | Authors: | Bucher, R.M, Mayans, O. | | Deposit date: | 2010-02-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the FnIII Tandem A77-A78 points to a periodically conserved architecture in the myosin-binding region of titin
J.Mol.Biol., 401, 2010
|
|
