6Q93
 
 | | MgADP-bound Fe protein of Vanadium nitrogenase from Azotobacter vinelandii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Rohde, M, Gerhardt, S, Einsle, O. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of VnfH, the iron protein component of vanadium nitrogenase.
J. Biol. Inorg. Chem., 23, 2018
|
|
6Q2W
 
 | | Crystal structure of human ROR gamma LBD in complex with a quinoline sulfonamide inverse agonist | | Descriptor: | (2~{S})-1-[2,4-bis(chloranyl)-3-[[4-imidazol-1-yl-2-(trifluoromethyl)quinolin-8-yl]oxymethyl]phenyl]sulfonyl-~{N}-methyl-pyrrolidine-2-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Ciesielski, F, Amaudrut, J, Argiriadi, M.A, Barth, M, Breinlinger, E.C, Calderwood, D.J, Cusack, K.P, Kort, M.E, Montalbetti, C, Potin, D, Poupardin, O, Spitzer, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of novel quinoline sulphonamide derivatives as potent, selective and orally active ROR gamma inverse agonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6QGK
 
 | | Structure of human Bcl-2 in complex with THIQ-phenyl pyrazole compound | | Descriptor: | 1-[2-[[(3~{S})-3-(aminomethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]phenyl]-~{N},~{N}-dibutyl-5-methyl-pyrazole-3-carboxamide, ACETATE ION, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QL4
 
 | | Crystal structure of nucleotide-free Mgm1 | | Descriptor: | 1,2-ETHANEDIOL, Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Wollweber, F, Pfitzner, A.-K, Muehleip, A, Sanchez, R, Kudryashev, M, Chiaruttin, N, Lilie, H, Schlegel, J, Rosenbaum, E, Hessenberger, M, Matthaeus, C, Noe, F, Roux, A, vanderLaan, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6PT3
 
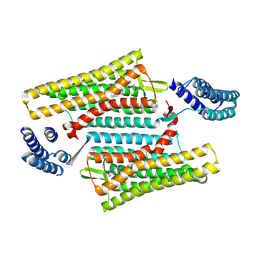 | | Crystal structure of the active delta opioid receptor in complex with the small molecule agonist DPI-287 | | Descriptor: | 4-[(R)-[(2S,5R)-4-benzyl-2,5-dimethylpiperazin-1-yl](3-hydroxyphenyl)methyl]-N,N-diethylbenzamide, Delta opioid receptor | | Authors: | Claff, T, Yu, J, Blais, V, Patel, N, Martin, C, Wu, L, Han, G.W, Holleran, B.J, Van der Poorten, O, Hanson, M.A, Sarret, P, Gendron, L, Cherezov, V, Katritch, V, Ballet, S, Liu, Z, Muller, C.E, Stevens, R.C. | | Deposit date: | 2019-07-14 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Sci Adv, 5, 2019
|
|
6Q5B
 
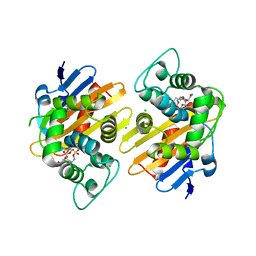 | | OXA-48_P68A-AVI. Evolutionary trade-offs of OXA-48 mediated ceftazidime-avibactam resistance | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, CARBON DIOXIDE, ... | | Authors: | Frohlich, C, Sorum, V, Thomassen, A.M, Johnsen, P.J, Leiros, H.K.S, Samuelsen, O. | | Deposit date: | 2018-12-07 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs.
Msphere, 4, 2019
|
|
6Q7J
 
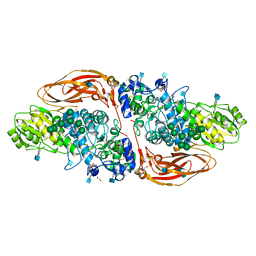 | | GH3 exo-beta-xylosidase (XlnD) in complex with xylobiose aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q5F
 
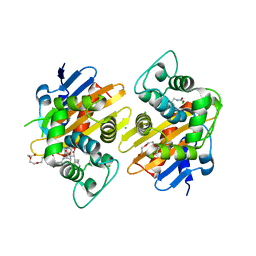 | | OXA-48_P68A-CAZ. Evolutionary trade-offs of OXA-48 mediated ceftazidime-avibactam resistance | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Frohlich, C, Sorum, V, Thomassen, A.M, Johnsen, P.J, Leiros, H.-K.S, Samuelsen, O. | | Deposit date: | 2018-12-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs.
Msphere, 4, 2019
|
|
6Q61
 
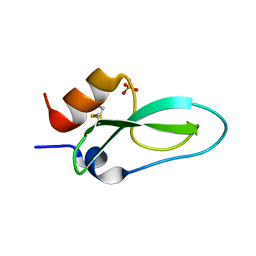 | | Pore-modulating toxins exploit inherent slow inactivation to block K+ channels | | Descriptor: | Kunitz-type conkunitzin-S1, SULFATE ION | | Authors: | Karbat, I, Gueta, H, Fine, S, Szanto, T, Hamer-Rogotner, S, Dym, O, Frolow, F, Gordon, D, Panyi, G, Gurevitz, M, Reuveny, E. | | Deposit date: | 2018-12-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pore-modulating toxins exploit inherent slow inactivation to block K+channels.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QB5
 
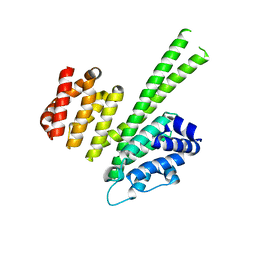 | | Crystal structure of the N-terminal region of human cohesin subunit STAG1 | | Descriptor: | Cohesin subunit SA-1, SODIUM ION | | Authors: | Newman, J.A, Katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-12-20 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
6QI9
 
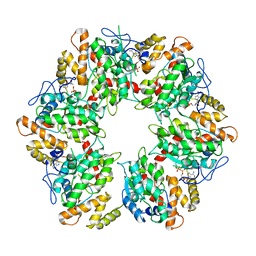 | | Truncated human R2TP complex, structure 4 (ADP-empty) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.63 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6Q7I
 
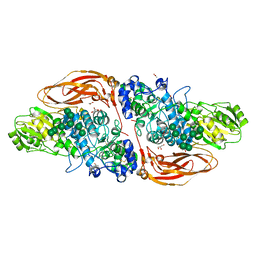 | | GH3 exo-beta-xylosidase (XlnD) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q8N
 
 | | GH10 endo-xylanase in complex with xylobiose epoxide inhibitor | | Descriptor: | (1~{R},2~{S},4~{S},5~{R})-cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6QI8
 
 | | Truncated human R2TP complex, structure 3 (ADP-filled) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6QFI
 
 | | Structure of human Mcl-1 in complex with BIM BH3 peptide | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QFQ
 
 | | Structure of human Mcl-1 in complex with indole acid inhibitor | | Descriptor: | 7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QG8
 
 | | Structure of human Bcl-2 in complex with PUMA BH3 peptide | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, Bcl-2-binding component 3 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QFM
 
 | | Structure of human Mcl-1 in complex with PUMA BH3 peptide | | Descriptor: | Bcl-2-binding component 3, CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGH
 
 | | Structure of human Bcl-2 in complex with ABT-263 | | Descriptor: | 4-(4-{[2-(4-chlorophenyl)-5,5-dimethylcyclohex-1-en-1-yl]methyl}piperazin-1-yl)-N-[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGJ
 
 | | Structure of human Bcl-2 in complex with fragment/ABT-263 hybrid | | Descriptor: | 4-[4-[(1~{R})-1-(6-methoxy-1,3-benzodioxol-5-yl)-2-pyrrolidin-1-yl-ethyl]phenyl]-~{N}-[4-[[(2~{R})-4-morpholin-4-yl-1-phenylsulfanyl-butan-2-yl]amino]-3-(trifluoromethylsulfonyl)phenyl]sulfonyl-benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QYL
 
 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QTY
 
 | |
6QU1
 
 | | Crystal structure of the KAP1 RBCC domain in complex with the SMARCAD1 CUE1 domain at 3.7 angstrom resolution. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1, Transcription intermediary factor 1-beta,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Newman, J.A, Aitkenhead, H, Gavard, A, Lim, M, Williams, H.L, Svejstrup, J.Q, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A Ubiquitin-Binding Domain that Binds a Structural Fold Distinct from that of Ubiquitin.
Structure, 2019
|
|
6RDI
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1A, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDX
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1F, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
