3KEB
 
 | | Thiol peroxidase from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, Probable thiol peroxidase, SULFATE ION | | Authors: | Osipiuk, J, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-25 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of thiol peroxidase from Chromobacterium violaceum
To be Published
|
|
5XIB
 
 | | Crystal Structure of HasAp with Fe-5,15-Diphenylporphyrin | | Descriptor: | 5,15-Diphenylporphyrin containing FE, Heme acquisition protein HasAp | | Authors: | Shoji, O, Uehara, H, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Heme Acquisition Protein HasA with Iron(III)-5,15-Diphenylporphyrin and Derivatives Thereof as an Artificial Prosthetic Group
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3KZ8
 
 | | Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs (p53-DNA complex 3) | | Descriptor: | Cellular tumor antigen p53, DNA (5'-D(*TP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*C)-3'), IODIDE ION, ... | | Authors: | Rozenberg, H, Suad, O, Shakked, Z. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KHL
 
 | | Dpo4 post-extension ternary complex with misinserted A opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2-AMINOFLUORENE, 5'-D(*CP*C*TP*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*AP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NKQ
 
 | | Crystal structure of mouse autotaxin in complex with 18:3-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E,12Z,15Z)-octadeca-9,12,15-trienoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
5XUZ
 
 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (CCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*CP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
1ENI
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENK
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
3NJW
 
 | |
3NI3
 
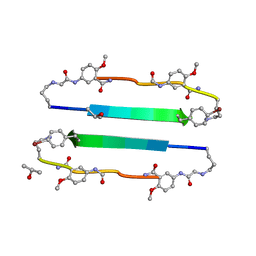 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
3N5M
 
 | | Crystals structure of a Bacillus anthracis aminotransferase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, CHLORIDE ION, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, DiLeo, R, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystals structure of a Bacillus anthracis aminotransferase
TO BE PUBLISHED
|
|
3NKO
 
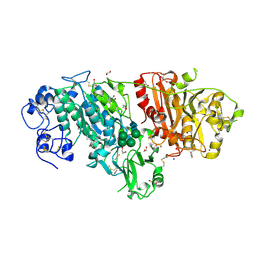 | | Crystal structure of mouse autotaxin in complex with 16:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NGH
 
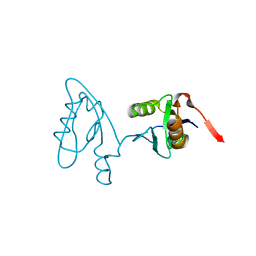 | | Molecular Analysis of the Interaction of the HDL Receptor SR-BI with the Adaptor Protein PDZK1 | | Descriptor: | PDZ domain-containing protein 1 | | Authors: | Kocher, O, Birrane, G, Krieger, M, Ladias, J.A. | | Deposit date: | 2010-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In vitro and in vivo analysis of the binding of the C terminus of the HDL receptor scavenger receptor class B, type I (SR-BI), to the PDZ1 domain of its adaptor protein PDZK1.
J.Biol.Chem., 285, 2010
|
|
3NKP
 
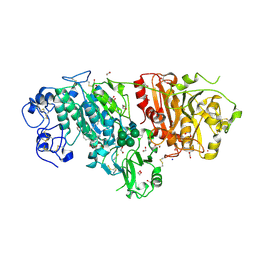 | | Crystal structure of mouse autotaxin in complex with 18:1-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKR
 
 | | Crystal structure of mouse autotaxin in complex with 22:6-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (4Z,7E,10E,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NGG
 
 | | X-ray Structure of Omwaprin | | Descriptor: | Omwaprin-a | | Authors: | Banigan, J.R, Mandal, K, Sawaya, M.R, Thammavongsa, V, Hendrickx, A.P.A, Schneewind, O, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Protein Sci., 19, 2010
|
|
3NX4
 
 | | Crystal structure of the yhdH oxidoreductase from Salmonella enterica in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Anderson, S.M, Wawrzak, Z, Peterson, S, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-12 | | Release date: | 2010-08-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the yhdH oxidoreductase from Salmonella enterica in complex with NADP
To be Published
|
|
3O04
 
 | | Crystal structure of the beta-keto-acyl carrier protein synthase II (lmo2201) from Listeria monocytogenes | | Descriptor: | beta-keto-acyl carrier protein synthase II | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the beta-keto-acyl carrier protein synthase II (lmo2201) from Listeria monocytogenes
To be Published
|
|
1E7U
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Perisic, O, Ried, C, Stephens, L, Williams, R.L. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1G5A
 
 | | AMYLOSUCRASE FROM NEISSERIA POLYSACCHAREA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMYLOSUCRASE, ... | | Authors: | Skov, L.K, Mirza, O, Henriksen, A, De Montalk, G.P, Remaud-Simeon, M, Sarcabal, P, Willemot, R.-M, Monsan, P, Gajhede, M. | | Deposit date: | 2000-10-31 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Amylosucrase, A Glucan-synthesizing Enzyme from the alpha-Amylase Family
J.Biol.Chem., 276, 2001
|
|
1FYM
 
 | |
1G3U
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE MONOPHOSPHATE (TMP) | | Descriptor: | MAGNESIUM ION, SULFATE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Li de la Sierra, I, Munier-Lehmann, H, Gilles, A.M, Barzu, O, Delarue, M. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray structure of TMP kinase from Mycobacterium tuberculosis complexed with TMP at 1.95 A resolution.
J.Mol.Biol., 311, 2001
|
|
1GAD
 
 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1FYL
 
 | |
1FYK
 
 | |
