9BD0
 
 | |
3E8X
 
 | | Putative NAD-dependent epimerase/dehydratase from Bacillus halodurans. | | 分子名称: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NAD-dependent epimerase/dehydratase | | 著者 | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-08-20 | | 公開日 | 2008-09-02 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-ray crystal structure of putative NAD-dependent epimerase/dehydratase from Bacillus halodurans.
To be Published
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1EGO
 
 | | NMR STRUCTURE OF OXIDIZED ESCHERICHIA COLI GLUTAREDOXIN: COMPARISON WITH REDUCED E. COLI GLUTAREDOXIN AND FUNCTIONALLY RELATED PROTEINS | | 分子名称: | GLUTAREDOXIN | | 著者 | Xia, T.-H, Bushweller, J.H, Sodano, P, Billeter, M, Bjornberg, O, Holmgren, A, Wuthrich, K. | | 登録日 | 1991-10-08 | | 公開日 | 1993-10-31 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
1VJ4
 
 | | SEQUENCE-DEPENDENT CONFORMATION OF AN A-DNA DOUBLE HELIX: THE CRYSTAL STRUCTURE OF THE OCTAMER D(G-G-T-A-T-A-C-C) | | 分子名称: | 5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3' | | 著者 | Shakked, Z, Rabinovich, D, Kennard, O, Cruse, W.B, Salisbury, S.A, Viswamitra, M.A. | | 登録日 | 1989-01-11 | | 公開日 | 1989-01-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Sequence-dependent conformation of an A-DNA double helix: The crystal structure of the octamer d(G-G-T-A-T-A-C-C)
J.Mol.Biol., 166, 1983
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | 分子名称: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3E7V
 
 | | Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-(3-aminophenyl)-N-(3-chlorophenyl)pyrazolo[1,5-a]pyrimidin-5-amine, NICKEL (II) ION, ... | | 著者 | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wickstroem, M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2008-08-19 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Human Haspin with a pyrazolo-pyrimidine ligand
To be Published
|
|
3ECY
 
 | |
3EDN
 
 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | 分子名称: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | 著者 | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-09-03 | | 公開日 | 2008-10-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
1DJB
 
 | |
3UXA
 
 | | Designed protein KE59 R1 7/10H | | 分子名称: | Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
9EMA
 
 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | 分子名称: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | 登録日 | 2024-03-07 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
9EMC
 
 | |
9BCZ
 
 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | 分子名称: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | 登録日 | 2024-04-10 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
8ZJF
 
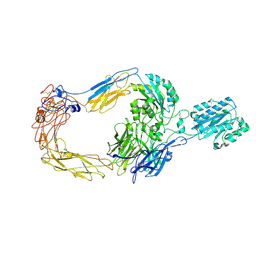 | | Cryo-EM structure of human integrin alpha-E beta-7 | | 分子名称: | CALCIUM ION, Integrin alpha-E, Integrin beta-7, ... | | 著者 | Akasaka, H, Nureki, O, Kise, Y. | | 登録日 | 2024-05-14 | | 公開日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structure of I domain-containing integrin alpha E beta 7.
Biochem.Biophys.Res.Commun., 721, 2024
|
|
3UZ5
 
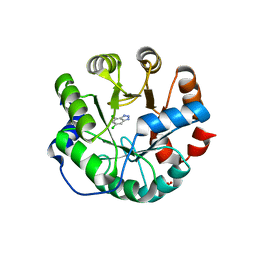 | | Designed protein KE59 R13 3/11H | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION, ... | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8YGJ
 
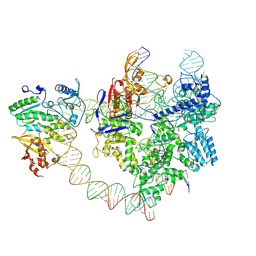 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2024-02-26 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
3UZJ
 
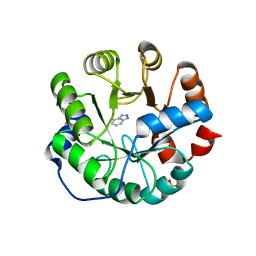 | | Designed protein KE59 R13 3/11H with benzotriazole | | 分子名称: | 1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY8
 
 | | Designed protein KE59 R5_11/5F | | 分子名称: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3E7Q
 
 | | The crystal structure of the putative transcriptional regulator from Pseudomonas aeruginosa PAO1 | | 分子名称: | transcriptional regulator | | 著者 | Zhang, R, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-08-18 | | 公開日 | 2008-10-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of the putative transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
6I5M
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDP and formate ion | | 分子名称: | FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Kravchenko, O, Nikonov, O, Gabdulkhakov, A, Stolboushkina, E, Arkhipova, V, Garber, M, Nikonov, S. | | 登録日 | 2018-11-13 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1EGR
 
 | | SEQUENCE-SPECIFIC 1H N.M.R. ASSIGNMENTS AND DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF REDUCED ESCHERICHIA COLI GLUTAREDOXIN | | 分子名称: | GLUTAREDOXIN | | 著者 | Sodano, P, Xia, T.-H, Bushweller, J.H, Bjornberg, O, Holmgren, A, Billeter, M, Wuthrich, K. | | 登録日 | 1991-10-08 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequence-specific 1H n.m.r. assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin.
J.Mol.Biol., 221, 1991
|
|
3ELK
 
 | | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum | | 分子名称: | CHLORIDE ION, Putative transcriptional regulator TA0346 | | 著者 | Grantz Saskova, K, Chruszcz, M, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-09-22 | | 公開日 | 2008-09-30 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum
To be Published
|
|
9F8X
 
 | |
8XGR
 
 | | ETB-eGt complex bound to endothelin-1 | | 分子名称: | Camelid antibody VHH fragment, Endothelin receptor type B, Endothelin-1, ... | | 著者 | Oshima, H.S, Sano, F.K, Akasaka, H, Iwama, A, Shihoya, W, Nureki, O. | | 登録日 | 2023-12-15 | | 公開日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Optimizing cryo-EM structural analysis of G i -coupling receptors via engineered G t and Nb35 application.
Biochem.Biophys.Res.Commun., 693, 2024
|
|
