3FT5
 
 | | Structure of HSP90 bound with a novel fragment | | Descriptor: | 4-methyl-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based Identification of Hsp90 Inhibitors.
Chemmedchem, 4, 2009
|
|
7ODU
 
 | | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D11, ... | | Authors: | Skalova, T, Blaha, J, Kalouskova, B, Skorepa, O, Vanek, O, Dohnalek, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11
To Be Published
|
|
6NW9
 
 | | CRYSTAL STRUCTURE OF A TAILSPIKE PROTEIN 3 (TSP3, ORF212) FROM ESCHERICHIA COLI O157:H7 BACTERIOPHAGE CBA120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J.Y, Herzberg, O. | | Deposit date: | 2019-02-06 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3).
Sci Rep, 9, 2019
|
|
1X8I
 
 | | Crystal Structure of the Zinc Carbapenemase CphA in Complex with the Antibiotic Biapenem | | Descriptor: | 5H-PYRAZOLO(1,2-A)(1,2,4)TRIAZOL-4-IUM, 6-((2-CARBOXY-6-(1-HYDROXYETHYL)-4-METHYL-7-OXO-1-AZABICYCLO(3.2.0)HEPT-2-EN-3-YL)THIO)-6,7-DIHYDRO-, HYDROXIDE, ... | | Authors: | Garau, G, Dideberg, O. | | Deposit date: | 2004-08-18 | | Release date: | 2004-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Metallo-beta-lactamase Enzyme in Action: Crystal Structures of the Monozinc Carbapenemase CphA and its Complex with Biapenem
J.Mol.Biol., 345, 2005
|
|
1X8G
 
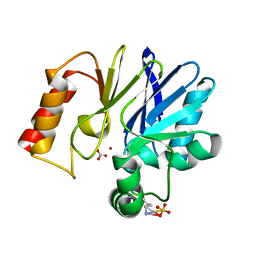 | |
1VTR
 
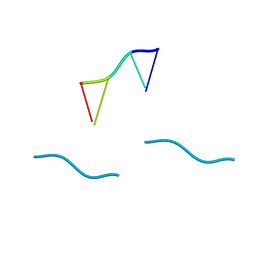 | | STRUCTURE OF THE DEOXYTETRANUCLEOTIDE D-PAPTPAPT AND A SEQUENCE-DEPENDENT MODEL FOR POLY(DA-DT) | | Descriptor: | DNA (5'-D(*AP*TP*AP*T)-3') | | Authors: | Viswamitra, M.A, Shakked, Z, Jones, P.G, Sheldrick, G.M, Salisbury, S.A, Kennard, O. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of the Deoxytetranucleotide d-pApTpApT and a Sequence-Dependent Model for Poly(dA-dT)
Biopolymers, 21, 1982
|
|
1ZDX
 
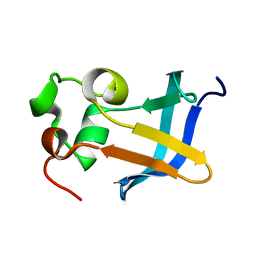 | | Solution Structure of the type 1 pilus assembly platform FimD(25-125) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
1ZDV
 
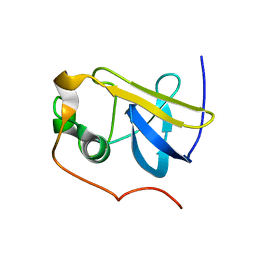 | | Solution Structure of the type 1 pilus assembly platform FimD(25-139) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
6W4Q
 
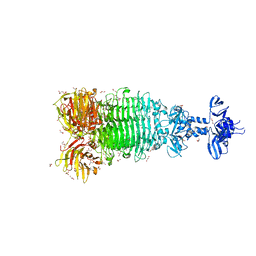 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J, Herzberg, O. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
3C0V
 
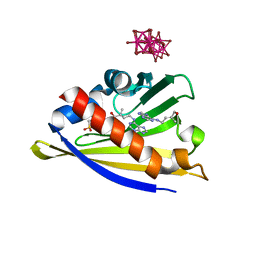 | | Crystal structure of cytokinin-specific binding protein in complex with cytokinin and Ta6Br12 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytokinin-specific binding protein, ... | | Authors: | Pasternak, O, Bujacz, A, Biesiadka, J, Bujacz, G, Sikorski, M, Jaskolski, M. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MAD phasing using the (Ta(6)Br(12))(2+) cluster: a retrospective study
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7PSQ
 
 | | Crystal structure of S100A4 labeled with NU074381b. | | Descriptor: | (2~{R},4~{R})-1-ethanoyl-~{N}-naphthalen-1-yl-4-phenyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Krojer, T, Arruda Bezerra, G, Koekemoer, L, Bountra, C, Von Delft, F, Brennan, P, Fedorov, O. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of S100A4 labeled with NU074381b.
To Be Published
|
|
3LJB
 
 | |
6HBD
 
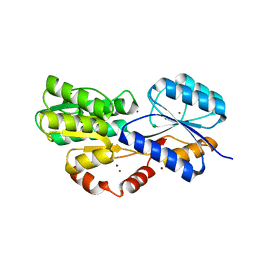 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Galactofuranose | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION, beta-D-galactofuranose | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6TVZ
 
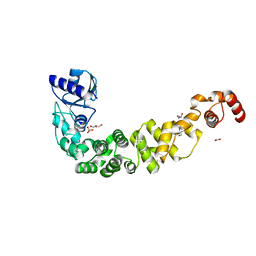 | | Structure of a psychrophilic CCA-adding enzyme crystallized in the XtalController device | | Descriptor: | ACETATE ION, CCA-adding enzyme, GLYCEROL, ... | | Authors: | de Wijn, R, Rollet, K, Coudray, L, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2020-01-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Monitoring the Production of High Diffraction-Quality Crystals of Two Enzymes in Real Time Using In Situ Dynamic Light Scattering
Crystals, 2020
|
|
1RXK
 
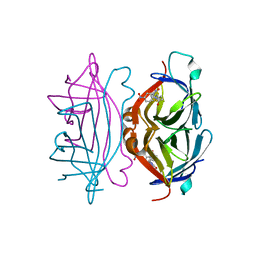 | | crystal structure of streptavidin mutant (M3) a combination of M1+M2 | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RXH
 
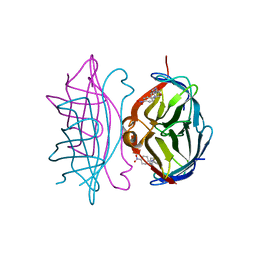 | | Crystal structure of streptavidin mutant L124R (M1) complexed with biotinyl p-nitroanilide (BNI) | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RP5
 
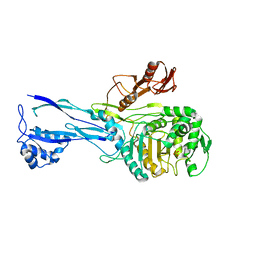 | | PBP2x from Streptococcus pneumoniae strain 5259 with reduced susceptibility to beta-lactam antibiotics | | Descriptor: | SULFATE ION, penicillin-binding protein 2x | | Authors: | Pernot, L, Chesnel, L, Legouellec, A, Croize, J, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics.
J.Biol.Chem., 279, 2004
|
|
3DAD
 
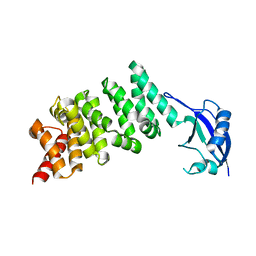 | | Crystal structure of the N-terminal regulatory domains of the formin FHOD1 | | Descriptor: | FH1/FH2 domain-containing protein 1 | | Authors: | Schulte, A, Stolp, B, Schonichen, A, Pylypenko, O, Rak, A, Fackler, O.T, Geyer, M. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Human Formin FHOD1 Contains a Bipartite Structure of FH3 and GTPase-Binding Domains Required for Activation.
Structure, 16, 2008
|
|
3JXV
 
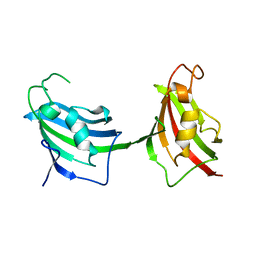 | |
1RXJ
 
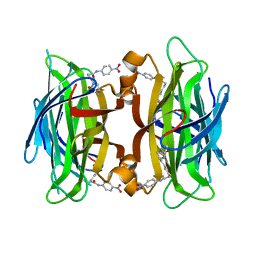 | | Crystal structure of streptavidin mutant (M2) where the L3,4 loop was replace by that of avidin | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3JPZ
 
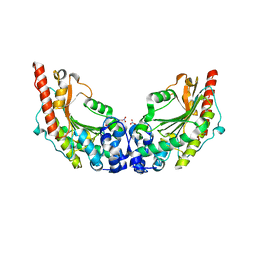 | | Crystal Structure of Lombricine Kinase | | Descriptor: | Lombricine kinase, NITRATE ION | | Authors: | Bush, D.J, Kirillova, O, Clark, S.A, Fabiola, F, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2009-09-04 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of lombricine kinase: implications for phosphagen kinase conformational changes.
J.Biol.Chem., 286, 2011
|
|
6HYH
 
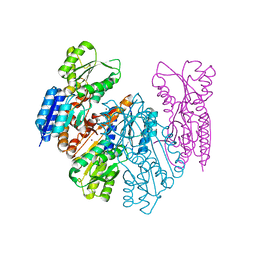 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Fucofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, ZINC ION, beta-D-fucofuranose | | Authors: | Li, M, Mueller, C, Zhang, L, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-10-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | Descriptor: | Transaldolase | | Authors: | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
6HB0
 
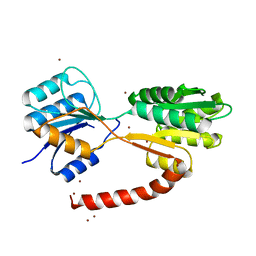 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
3LMB
 
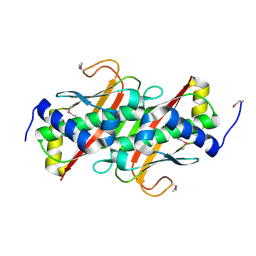 | | The crystal structure of the protein OLEI01261 with unknown function from Chlorobaculum tepidum TLS | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
