1GLN
 
 | | ARCHITECTURES OF CLASS-DEFINING AND SPECIFIC DOMAINS OF GLUTAMYL-TRNA SYNTHETASE | | Descriptor: | GLUTAMYL-TRNA SYNTHETASE | | Authors: | Nureki, O, Vassylyev, D.G, Katayanagi, K, Shimizu, T, Sekine, S, Kigawa, T, Miyazawa, T, Yokoyama, S, Morikawa, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1994-07-20 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architectures of class-defining and specific domains of glutamyl-tRNA synthetase.
Science, 267, 1995
|
|
1UEV
 
 | |
1UET
 
 | |
1UEU
 
 | |
1V2X
 
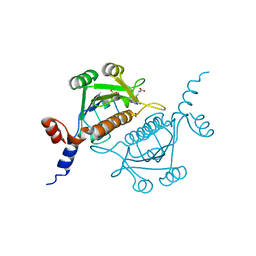 | | TrmH | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (Gm18) methyltransferase | | Authors: | Nureki, O, Watanabe, K, Fukai, S, Ishii, R, Endo, Y, Hori, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-17 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deep Knot Structure for Construction of Active Site and Cofactor Binding Site of tRNA Modification Enzyme
STRUCTURE, 12, 2004
|
|
3AII
 
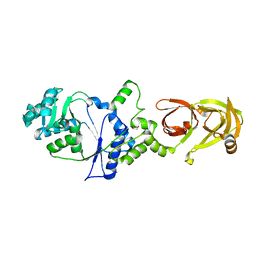 | |
2ZY9
 
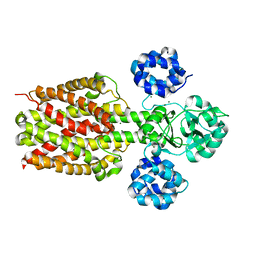 | |
2D6F
 
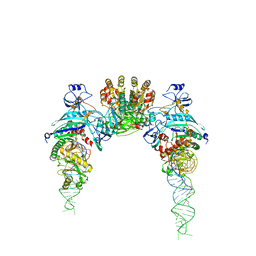 | |
1IRX
 
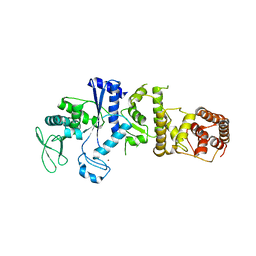 | | Crystal structure of class I lysyl-tRNA synthetase | | Descriptor: | ZINC ION, lysyl-tRNA synthetase | | Authors: | Nureki, O, Terada, T, Ishitani, R, Ambrogelly, A, Ibba, M, Soll, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-10-25 | | Release date: | 2002-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional convergence of two lysyl-tRNA synthetases with unrelated topologies.
Nat.Struct.Biol., 9, 2002
|
|
1ILE
 
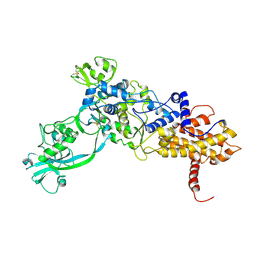 | | ISOLEUCYL-TRNA SYNTHETASE | | Descriptor: | ISOLEUCYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Nureki, O, Vassylyev, D.G, Tateno, M, Shimada, A, Nakama, T, Fukai, S, Konno, M, Schimmel, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-02-24 | | Release date: | 1999-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme structure with two catalytic sites for double-sieve selection of substrate.
Science, 280, 1998
|
|
1IPA
 
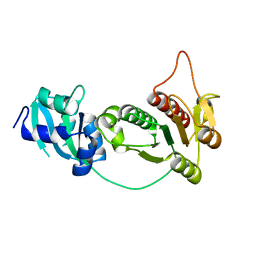 | | CRYSTAL STRUCTURE OF RNA 2'-O RIBOSE METHYLTRANSFERASE | | Descriptor: | RNA 2'-O-RIBOSE METHYLTRANSFERASE | | Authors: | Nureki, O, Shirouzu, M, Hashimoto, K, Ishitani, R, Terada, T, Tamakoshi, M, Oshima, T, Chijimatsu, M, Takio, K, Vassylyev, D.G, Shibata, T, Inoue, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An enzyme with a deep trefoil knot for the active-site architecture.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7XJI
 
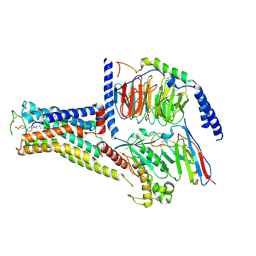 | | Solabegron-activated dog beta3 adrenergic receptor | | Descriptor: | 3-[3-[2-[[(2~{S})-2-(3-chlorophenyl)-2-oxidanyl-ethyl]amino]ethylamino]phenyl]benzoic acid, Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the beta 3 adrenergic receptor bound to solabegron and isoproterenol.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
7XJH
 
 | | Isoproterenol-activated dog beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
8ZH8
 
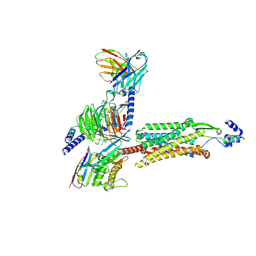 | | Human GPR103 -Gq complex bound to QRFP26 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-2,Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Iwama, A, Akasaka, H, Sano, F.K, Oshima, H.S, Shihoya, W, Nureki, O. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure and dynamics of the pyroglutamylated RF-amide peptide QRFP receptor GPR103.
Nat Commun, 15, 2024
|
|
8YNY
 
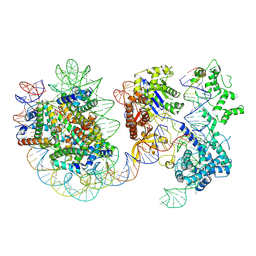 | | Structure of Cas9-sgRNA ribonucleoprotein bound to nucleosome | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (17-mer), DNA (175-mer), ... | | Authors: | Nagamura, R, Kujirai, T, Kusakizako, T, Hirano, H, Kurumizaka, H, Nureki, O. | | Deposit date: | 2024-03-12 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Structural insights into how Cas9 targets nucleosomes.
Nat Commun, 15, 2024
|
|
4WIB
 
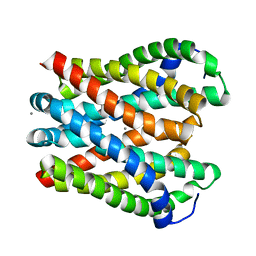 | | Crystal structure of Magnesium transporter MgtE | | Descriptor: | CALCIUM ION, Magnesium transporter MgtE | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-09-25 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
9J5V
 
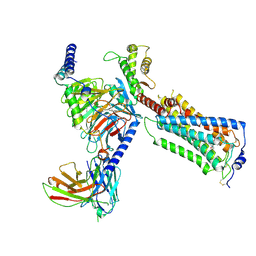 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to CpY | | Descriptor: | CpY, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2024-08-13 | | Release date: | 2024-11-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural mechanisms of potent lysophosphatidic acid receptor 1 activation by nonlipid basic agonists.
Commun Biol, 7, 2024
|
|
4GEN
 
 | | Crystal structure of Zucchini (monomer) | | Descriptor: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4GEM
 
 | | Crystal structure of Zucchini (K171A) | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5L
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the absence of NADH (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5K
 
 | | Time-resolved SFX structure of cytochrome P450nor : 20 ms after photo-irradiation of caged NO in the absence of NADH (NO-bound state), light data | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
2RRN
 
 | | Solution structure of SecDF periplasmic domain P4 | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tanaka, T, Tsukazaki, T, Echizen, Y, Nureki, O, Kohno, T. | | Deposit date: | 2011-01-30 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
