7O0G
 
 | |
7O0H
 
 | |
8R41
 
 | | Structure of CHI3L1 in complex with inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
1YS6
 
 | |
1YSR
 
 | |
1YS3
 
 | |
1YS7
 
 | | Crystal structure of the response regulator protein prrA complexed with Mg2+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The structural basis of signal transduction for the response regulator PrrA from Mycobacterium tuberculosis.
J.Biol.Chem., 281, 2006
|
|
4HKQ
 
 | | XMRV reverse transcriptase in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*TP*GP*GP*AP*AP*TP*CP*A*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*TP*CP*TP*G)-3'), RNA (5'-R(*AP*AP*CP*AP*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*CP*AP*U)-3'), Reverse transcriptase/ribonuclease H p80 | | Authors: | Nowak, E, Potrzebowski, W, Konarev, P.V, Rausch, J.W, Bona, M.K, Svergun, D.I, Bujnicki, J.M, Le Grice, S.F.J, Nowotny, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid
Nucleic Acids Res., 41, 2013
|
|
8R42
 
 | | Structure of CHI3L1 in complex with inhibititor 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2~{R})-2-[(4-chlorophenyl)methyl]pyrrolidin-1-yl]piperidin-1-yl]pyridine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
8R4X
 
 | | Structure of Chitinase-3-like protein 1 in complex with inhibitor 30 | | Descriptor: | (2~{S},5~{S})-4-[1-(4-chloranylpyridin-2-yl)piperidin-4-yl]-5-[(4-chlorophenyl)methyl]-2-methyl-morpholine, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
1MNZ
 
 | | Atomic structure of Glucose isomerase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P.A. | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Atomic structure of Glucose isomerase
To be published
|
|
1O1H
 
 | | STRUCTURE OF GLUCOSE ISOMERASE DERIVATIZED WITH KR. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P.A. | | Deposit date: | 2002-11-07 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Glucose Isomerase Derivatized with Kr.
To be Published
|
|
4OL8
 
 | | Ty3 reverse transcriptase bound to DNA/RNA | | Descriptor: | 5'-D(*CP*AP*TP*CP*TP*TP*CP*CP*TP*CP*TP*CP*TP*CP*TP*C)-3', 5'-R(*CP*UP*GP*AP*GP*AP*GP*AP*GP*AP*GP*GP*AP*AP*GP*AP*UP*G)-3', Reverse transcriptase/ribonuclease H, ... | | Authors: | Nowak, E, Miller, J.T, Bona, M.K, Studnicka, J, Szczepanowski, R.H, Jurkowski, J, Le Grice, S.F.J, Nowotny, M. | | Deposit date: | 2014-01-23 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8AUP
 
 | | Structure of hARG1 with a novel inhibitor. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[(1~{R},3~{R},4~{S})-3-azanyl-3-carboxy-4-[(dimethylamino)methyl]cyclohexyl]ethyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, ... | | Authors: | Napiorkowska-Gromadzka, A, Nowak, E, Nowotny, M. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Arginase 1/2 Inhibitor OATD-02: From Discovery to First-in-man Setup in Cancer Immunotherapy.
Mol.Cancer Ther., 22, 2023
|
|
6F4A
 
 | | Yeast mitochondrial RNA degradosome complex mtEXO | | Descriptor: | Exoribonuclease II, mitochondrial, RNA (5'-R(P*AP*GP*AP*UP*AP*C)-3'), ... | | Authors: | Razew, M, Nowak, E, Nowotny, M. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components.
Nat Commun, 9, 2018
|
|
5J5Y
 
 | | Translation initiation factor 4E in complex with m2(7,2'O)GppCCl2ppG mRNA 5' cap analog | | Descriptor: | 2-amino-9-{5-O-[(R)-{[(S)-{dichloro[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2-O-methyl-beta-D-ribofuranosyl}-7-methyl-9H-purin-7-ium-6-olate, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
3HP4
 
 | | Crystal structure of psychrotrophic esterase EstA from Pseudoalteromonas sp. 643A inhibited by monoethylphosphonate | | Descriptor: | GDSL-esterase | | Authors: | Brzuszkiewicz, A, Nowak, E, Dauter, Z, Dauter, M, Cieslinski, H, Kur, J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of EstA esterase from psychrotrophic Pseudoalteromonas sp. 643A covalently inhibited by monoethylphosphonate.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5J5O
 
 | | Translation initiation factor 4E in complex with m7GppppG mRNA 5' cap analog | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]-7-methylguanosine, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
8BDC
 
 | | Human apo TRPM8 in a closed state (composite map) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Palchevskyi, S, Czarnocki-Cieciura, M, Vistoli, G, Gervasoni, S, Nowak, E, Beccari, A.R, Nowotny, M, Talarico, C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structure of human TRPM8 channel.
Commun Biol, 6, 2023
|
|
6F3H
 
 | | Crystal structure of Dss1 exoribonuclease active site mutant D477N from Candida glabrata | | Descriptor: | Exoribonuclease II, mitochondrial, MAGNESIUM ION, ... | | Authors: | Razew, M, Nowak, E, Nowotny, M. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components.
Nat Commun, 9, 2018
|
|
8R0S
 
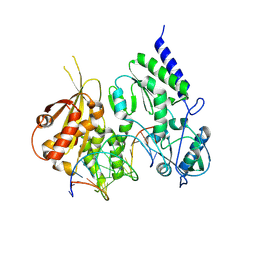 | | Structure of reverse transcriptase from Cauliflower Mosaic Virus in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*CP*GP*CP*AP*CP*TP*GP*CP*TP*GP*GP*A)-3'), Enzymatic polyprotein, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*AP*GP*UP*GP*CP*GP*UP*AP*GP*C)-3') | | Authors: | Prabaharan, C, Figiel, M, Chamera, S, Szczepanowski, R, Nowak, E, Nowotny, M. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical characterization of cauliflower mosaic virus reverse transcriptase.
J.Biol.Chem., 300, 2024
|
|
6GKK
 
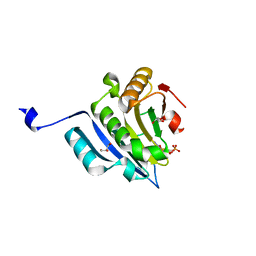 | | Translation initiation factor 4E in complex with beta-phosphorothioate trinucleotide mRNA 5' cap diastereomer 1 (m7GppSpApG D1) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl [phosphonooxy(sulfanyl)phosphoryl] hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Translation initiation factor 4E in complex with beta-phosphorothioate trinucleotide mRNA 5' cap diastereomer 1 (m7GppSpApG D1)
To Be Published
|
|
6GKJ
 
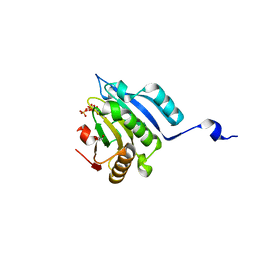 | | Translation initiation factor 4E in complex with trinucleotide mRNA 5' cap (m7GpppApG) | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Translation initiation factor 4E in complex with trinucleotide mRNA 5' cap (m7GpppApG)
To Be Published
|
|
6GKL
 
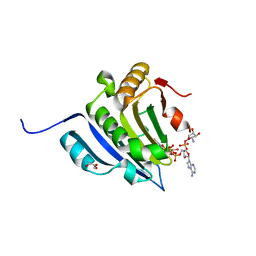 | | Translation initiation factor 4E in complex with beta-phosphorothioate trinucleotide mRNA 5' cap diastereomer 2 (m7GppSpApG D2) | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-azanyl-4,5-dihydropurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Translation initiation factor 4E in complex with beta-phosphorothioate trinucleotide mRNA 5' cap diastereomer 2 (m7GppSpApG D2)
To Be Published
|
|
8QKF
 
 | | SmtB protein of Mycobacterium smegmatis | | Descriptor: | ArsR family transcriptional regulator | | Authors: | Potocki, S, Pyra, A, Nowak, E, Rola, A, Plaskowka, K, Trojanowski, D, Holowka, J, Pasikowski, P, Gumienna-Kontecka, E, Zakrzewska-Czerwinska, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | SmtB protein of Mycobacterium smegmatis
To Be Published
|
|
