3A7P
 
 | |
3A77
 
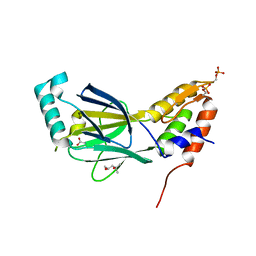 | | The crystal structure of phosphorylated IRF-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Interferon regulatory factor 3 | | Authors: | Takahasi, K, Horiuchi, M, Noda, N.N, Inagaki, F. | | Deposit date: | 2009-09-17 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change.
Genes Cells, 15, 2010
|
|
3A7O
 
 | |
2ZZP
 
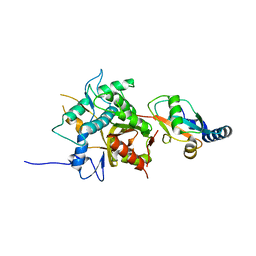 | |
3VGP
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AF_0329) from Archaeoglobus fulgidus | | Descriptor: | Transmembrane oligosaccharyl transferase, putative | | Authors: | Matsumoto, S, Igura, M, Nyirenda, J, Yuzawa, S, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the C-Terminal Globular Domain of Oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 A Resolution
Biochemistry, 51, 2012
|
|
3VU0
 
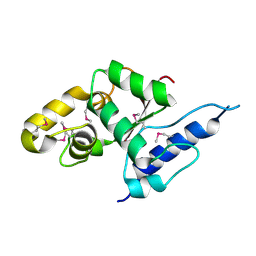
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AfAglB-S2, AF_0040, O30195_ARCFU) from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
3VU4
 
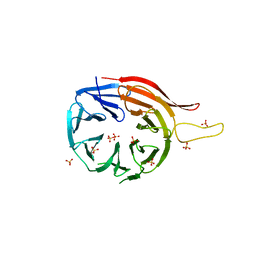 | |
3VGO
 
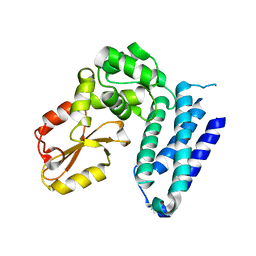 | |
3VX7
 
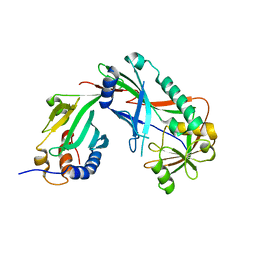 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VU1
 
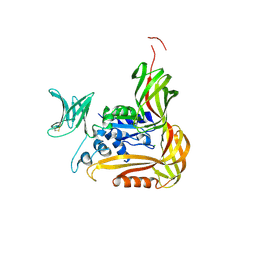 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PhAglB-L, O74088_PYRHO) from Pyrococcus horikoshii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein PH0242 | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
3VX6
 
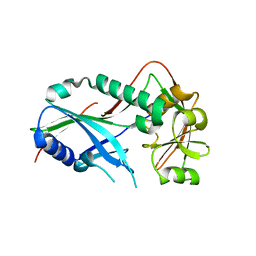 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VQI
 
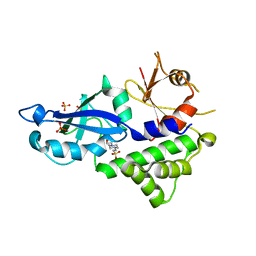 | | Crystal structure of Kluyveromyces marxianus Atg5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Atg5, SULFATE ION | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-03-24 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
