7CMA
 
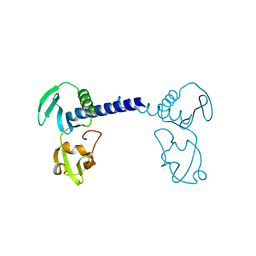 | | Structure of A151R from African swine fever virus Georgia | | 分子名称: | A151R, ZINC ION | | 著者 | Niu, D, Liu, K, Huang, J, Chen, C, Liu, W, Guo, R. | | 登録日 | 2020-07-26 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure basis of non-structural protein pA151R from African Swine Fever Virus.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
7VVC
 
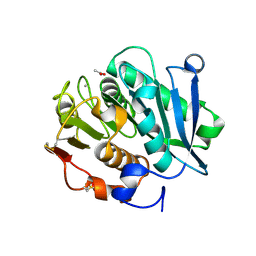 | | Crystal structure of inactive mutant of leaf-branch compost cutinase variant | | 分子名称: | ACETATE ION, ACETIC ACID, CALCIUM ION, ... | | 著者 | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | 登録日 | 2021-11-05 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W45
 
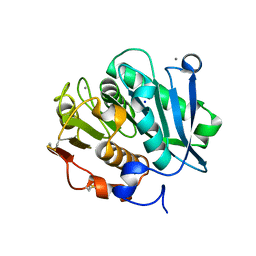 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | 分子名称: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | 著者 | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | 登録日 | 2021-11-26 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W44
 
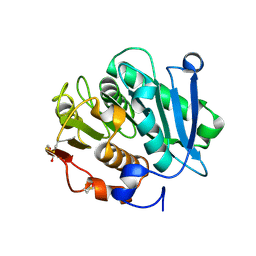 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | 分子名称: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | 著者 | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | 登録日 | 2021-11-26 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7VVE
 
 | | Complex structure of a leaf-branch compost cutinase variant in complex with mono(2-hydroxyethyl) terephthalic acid | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, ... | | 著者 | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | 登録日 | 2021-11-05 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W1N
 
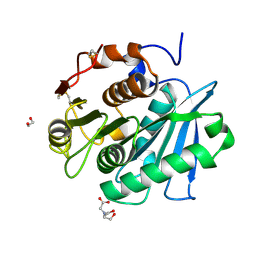 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | 分子名称: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | 著者 | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
3ZIM
 
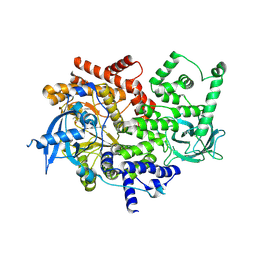 | | Discovery of a potent and isoform-selective targeted covalent inhibitor of the lipid kinase PI3Kalpha | | 分子名称: | 1-[4-[[2-(1H-indazol-4-yl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidin-6-yl]methyl]piperazin-1-yl]-6-methyl-hept-5-ene-1,4- dione, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | 著者 | Nacht, M, Qiao, L, Sheets, M.P, Martin, T.S, Labenski, M, Mazdiyasni, H, Karp, R, Zhu, Z, Chaturvedi, P, Bhavsar, D, Niu, D, Westlin, W, Petter, R.C, Medikonda, A.P, Jestel, A, Blaesse, M, Singh, J. | | 登録日 | 2013-01-09 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Discovery of a Potent and Isoform-Selective Targeted Covalent Inhibitor of the Lipid Kinase Pi3Kalpha
J.Med.Chem., 56, 2013
|
|
3OYP
 
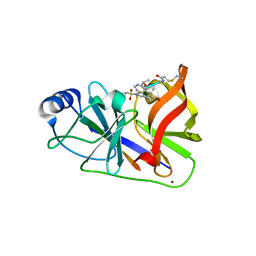 | | HCV NS3/4A in complex with ligand 3 | | 分子名称: | Non-structural protein 4A, Peptidomimetic inhibitor, Serine protease NS3, ... | | 著者 | Hagel, M, Niu, D, St.Martin, T, Sheets, M.P, Qiao, L, Bernard, H, Karp, R.M, Zhu, Z, Labenski, M.T, Chaturvedi, P.C, Nacht, M, Westlin, W.F, Petter, R.C, Singh, J. | | 登録日 | 2010-09-23 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Selective irreversible inhibition of a protease by targeting a noncatalytic cysteine.
Nat.Chem.Biol., 7, 2011
|
|
8J45
 
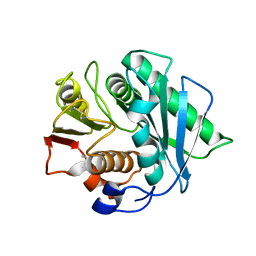 | | Crystal structure of a Pichia pastoris-expressed IsPETase variant | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly(ethylene terephthalate) hydrolase | | 著者 | Li, X, He, H.L, Long, X, Niu, D, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-04-19 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Complete decomposition of poly(ethylene terephthalate) by crude PET hydrolytic enzyme produced in Pichia pastoris
Chem Eng J, 2023
|
|
8IHR
 
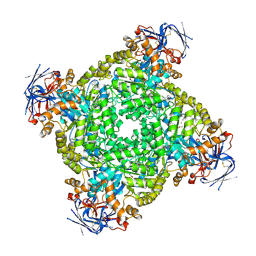 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | 分子名称: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | 分子名称: | Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8J85
 
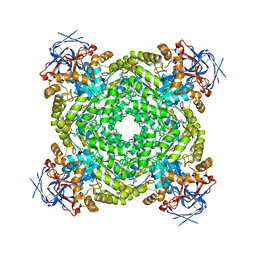 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-04-30 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
