2W9Q
 
 | | Crystal Structure of Potato Multicystatin-P212121 | | Descriptor: | MULTICYSTATIN | | Authors: | Nissen, M.S, Kumar, G.N, Youn, B, Knowles, D.B, Lam, K.S, Ballinger, W.J, Knowles, N.R, Kang, C. | | Deposit date: | 2009-01-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of Solanum Tuberosum Multicystatin and its Structural Comparison with Other Cystatins.
Plant Cell, 21, 2009
|
|
2W9P
 
 | | Crystal Structure of Potato Multicystatin | | Descriptor: | MULTICYSTATIN | | Authors: | Nissen, M.S, Kumar, G.N, Youn, B, Knowles, D.B, Lam, K.S, Ballinger, W.J, Knowles, N.R, Kang, C. | | Deposit date: | 2009-01-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Solanum Tuberosum Multicystatin and its Structural Comparison with Other Cystatins.
Plant Cell, 21, 2009
|
|
4LTM
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
4LZI
 
 | | Characterization of Solanum tuberosum Multicystatin and Significance of Core Domains | | Descriptor: | Multicystatin | | Authors: | Nissen, M.S, Kumar, G.N, Green, A.R, Knowles, N.R, Kang, C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Solanum tuberosum Multicystatin and the Significance of Core Domains.
Plant Cell, 25, 2013
|
|
4LTD
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - apo form | | Descriptor: | NADH-dependent FMN reductase, PHOSPHATE ION, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
4LTN
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN, NADH complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, ... | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
4HUZ
 
 | | 2,6-Dichloro-p-hydroquinone 1,2-Dioxygenase | | Descriptor: | 2,6-dichloro-p-hydroquinone 1,2-dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Hayes, R.P, Nissen, M.S, Green, A.R, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2012-11-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) from Sphingobium chlorophenolicum, a new type of aromatic ring-cleavage enzyme.
Mol.Microbiol., 88, 2013
|
|
4G5E
 
 | | 2,4,6-Trichlorophenol 4-monooxygenase | | Descriptor: | 2,4,6-Trichlorophenol 4-monooxygenase | | Authors: | Hayes, R.P, Webb, B.N, Subramanian, A.K, Nissen, M, Popchock, A, Xun, L, Kang, C. | | Deposit date: | 2012-07-17 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Catalytic Differences between Two FADH(2)-Dependent Monooxygenases: 2,4,5-TCP 4-Monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6-TCP 4-Monooxygenase (TcpA) from Cupriavidus necator JMP134.
Int J Mol Sci, 13, 2012
|
|
3S2I
 
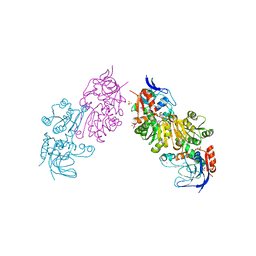 | | Crystal Structure of FurX NADH+:Furfuryl alcohol II | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3S1L
 
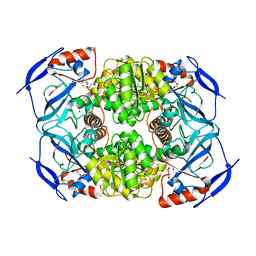 | | Crystal Structure of Apo-form FurX | | Descriptor: | HEXAETHYLENE GLYCOL, ZINC ION, Zinc-containing alcohol dehydrogenase superfamily | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-15 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Furfural reduction mechanism of a zinc-dependent alcohol dehydrogenase from Cupriavidus necator JMP134.
Mol.Microbiol., 83, 2012
|
|
3S2E
 
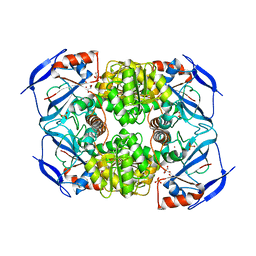 | | Crystal Structure of FurX NADH Complex 1 | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal Structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3S2F
 
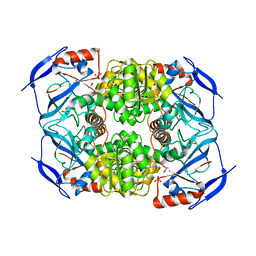 | | Crystal Structure of FurX NADH:Furfural | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHORYLISOPROPANE, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3S2G
 
 | | Crystal Structure of FurX NADH+:Furfuryl alcohol I | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3TRQ
 
 | | Crystal structure of native rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Glycosylation of Skeletal Calsequestrin: IMPLICATIONS FOR ITS FUNCTION.
J.Biol.Chem., 287, 2012
|
|
3US3
 
 | | Recombinant rabbit skeletal calsequestrin-MPD complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Nissen, M.S, Munske, G.R, Kang, C. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Glycosylation of skeletal calsequestrin: implications for its function.
J.Biol.Chem., 287, 2012
|
|
4NPX
 
 | | Structure of hypothetical protein Cj0539 from Campylobacter jejuni | | Descriptor: | Putative uncharacterized protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Adkins, J.N, Endres, M, Nissen, M, Konkel, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-11-22 | | Release date: | 2014-01-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of hypothetical protein Cj0539 from Campylobacter jejuni
To be Published
|
|
3TRP
 
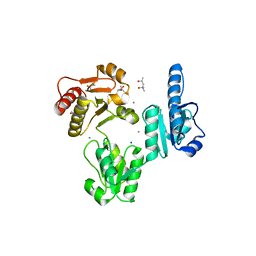 | | Crystal structure of recombinant rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8817 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
3UOM
 
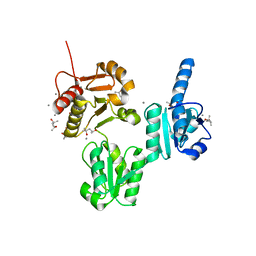 | | Ca2+ complex of Human skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Danna, B.R, Nissen, M.S, Kang, C.H. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
2R59
 
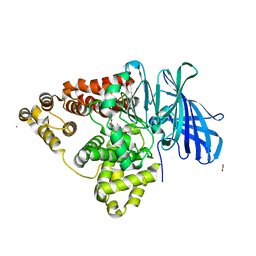 | | Leukotriene A4 hydrolase complexed with inhibitor RB3041 | | Descriptor: | ACETIC ACID, Leukotriene A-4 hydrolase, N-{(2S)-3-[(R)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Tholander, F, Haeggstrom, J.Z, Thunnissen, M, Muroya, A, Roques, B.P, Fournie-Zaluski, M.C. | | Deposit date: | 2007-09-03 | | Release date: | 2008-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
3B7R
 
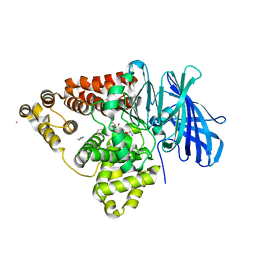 | | Leukotriene A4 Hydrolase Complexed with Inhibitor RB3040 | | Descriptor: | IMIDAZOLE, Leukotriene A-4 hydrolase, N-[3-[(1-AMINOETHYL)(HYDROXY)PHOSPHORYL]-2-(1,1'-BIPHENYL-4-YLMETHYL)PROPANOYL]ALANINE, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
3B7S
 
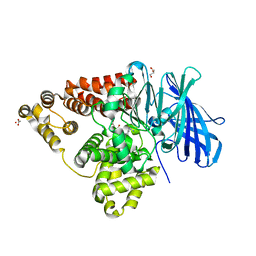 | | [E296Q]LTA4H in complex with RSR substrate | | Descriptor: | ACETIC ACID, GLYCEROL, Leukotriene A-4 hydrolase, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.465 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
3B7U
 
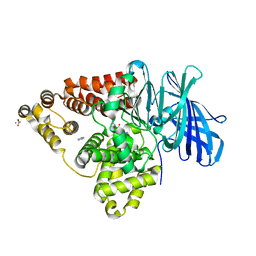 | | Leukotriene A4 Hydrolase Complexed with KELatorphan | | Descriptor: | ACETIC ACID, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
3B7T
 
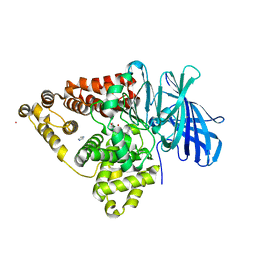 | | [E296Q]LTA4H in complex with Arg-Ala-Arg substrate | | Descriptor: | IMIDAZOLE, Leukotriene A-4 hydrolase, RAR peptide, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
1LO5
 
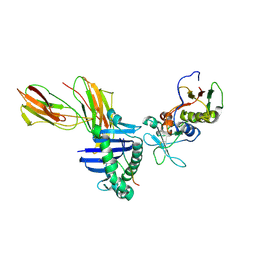 | | Crystal structure of the D227A variant of Staphylococcal enterotoxin A in complex with human MHC class II | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DR-1 beta chain, ... | | Authors: | Petersson, K, Thunnissen, M, Forsberg, G, Walse, B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a SEA Variant in Complex with MHC Class II Reveals the Ability of SEA to Crosslink MHC Molecules
Structure, 10, 2002
|
|
2UWF
 
 | | Crystal structure of family 10 xylanase from Bacillus halodurans | | Descriptor: | ALKALINE ACTIVE ENDOXYLANASE, CALCIUM ION, COPPER (II) ION | | Authors: | Mamo, G, Thunnissen, M, Hatti-Kaul, R, Mattiasson, B. | | Deposit date: | 2007-03-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Alkaline Active Xylanase: Insights Into Mechanisms of High Ph Catalytic Adaptation
Biochimie, 91, 2009
|
|
