3VM6
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1 in complex with alpha-D-ribose-1,5-bisphosphate | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, A, Fujihashi, M, Aono, R, Sato, T, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2011-12-08 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
3VS8
 
 | | Crystal structure of type III PKS ArsC | | Descriptor: | SODIUM ION, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
3VGM
 
 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose | | Descriptor: | Glucokinase, POTASSIUM ION, ZINC ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
1Y1C
 
 | | Solution structure of Anemonia elastase inhibitor analogue | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
3VGL
 
 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose and AMPPNP | | Descriptor: | Glucokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
7CMN
 
 | |
5Y52
 
 | |
1D8J
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
3W6V
 
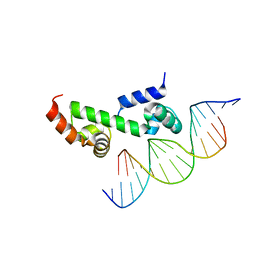 | | Crystal structure of the DNA-binding domain of AdpA, the global transcriptional factor, in complex with a target DNA | | Descriptor: | AdpA, DNA (5'-D(*AP*GP*GP*TP*TP*GP*GP*CP*GP*GP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*TP*GP*TP*GP*AP*AP*CP*CP*CP*GP*CP*CP*AP*AP*C)-3') | | Authors: | Yao, M.D, Ohtsuka, J, Nagata, K, Miyazono, K, Ohnishi, Y, Tanokura, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Complex Structure of the DNA-binding Domain of AdpA, the Global Transcription Factor in Streptomyces griseus, and a Target Duplex DNA Reveals the Structural Basis of Its Tolerant DNA Sequence Specificity
J.Biol.Chem., 288, 2013
|
|
1Y1B
 
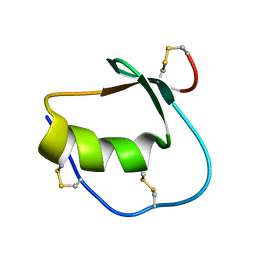 | | Solution structure of Anemonia elastase inhibitor | | Descriptor: | Elastase inhibitor | | Authors: | Hemmi, H, Kumazaki, T, Yoshizawa-Kumagaye, K, Nishiuchi, Y, Yoshida, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2004-11-18 | | Release date: | 2005-07-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Study of an Anemonia Elastase Inhibitor, a "Nonclassical" Kazal-Type Inhibitor from Anemonia sulcata
Biochemistry, 44, 2005
|
|
3A9C
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakaraensis KOD1 in complex with ribulose-1,5-bisphosphate | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, RIBULOSE-1,5-DIPHOSPHATE, ... | | Authors: | Nakamura, A, Fujihashi, M, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
3WP9
 
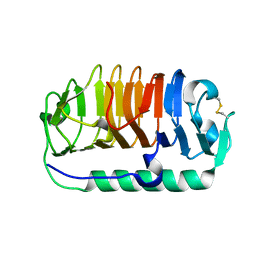 | | Crystal structure of antifreeze protein from an Antarctic sea ice bacterium Colwellia sp. | | Descriptor: | Ice-binding protein | | Authors: | Hanada, Y, Nishimiya, Y, Miura, A, Tsuda, S, Kondo, H. | | Deposit date: | 2014-01-10 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hyperactive antifreeze protein from an Antarctic sea ice bacterium Colwellia sp. has a compound ice-binding site without repetitive sequences.
Febs J., 281, 2014
|
|
3WQP
 
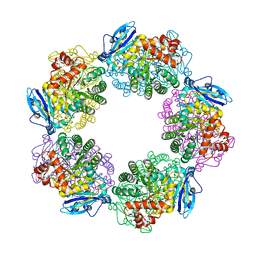 | | Crystal structure of Rubisco T289D mutant from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujihashi, M, Nishitani, Y, Kiriyama, T, Miki, K. | | Deposit date: | 2014-01-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutation design of thermophilic Rubisco based on the three-dimensional structure enhances its activity at ambient temperature
to be published
|
|
1VD4
 
 | | Solution structure of the zinc finger domain of TFIIE alpha | | Descriptor: | Transcription initiation factor IIE, alpha subunit, ZINC ION | | Authors: | Okuda, M, Tanaka, A, Arai, Y, Satoh, M, Okamura, H, Nagadoi, A, Hanaoka, F, Ohkuma, Y, Nishimura, Y. | | Deposit date: | 2004-03-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc finger structure in the large subunit of human general transcription factor TFIIE.
J.Biol.Chem., 279, 2004
|
|
1ETL
 
 | |
3VS9
 
 | | Crystal structure of type III PKS ArsC mutant | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
1ETM
 
 | |
6KXD
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
5XNZ
 
 | | Crystal structure of CreD complex with fumarate | | Descriptor: | CreD, FUMARIC ACID | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
5XNY
 
 | | Crystal structure of CreD | | Descriptor: | CreD | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
1ETN
 
 | |
7FBH
 
 | | geranyl pyrophosphate C6-methyltransferase BezA | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BezA, ... | | Authors: | Tsutsumi, H, Moriwaki, Y, Terada, T, Shimizu, K, Katsuyama, Y, Ohnishi, Y. | | Deposit date: | 2021-07-10 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Molecular Basis of the Catalytic Mechanism of Geranyl Pyrophosphate C6-Methyltransferase: Creation of an Unprecedented Farnesyl Pyrophosphate C6-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7FBO
 
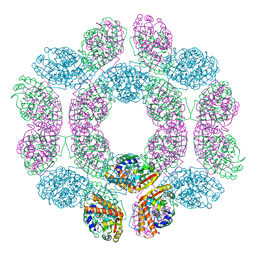 | | geranyl pyrophosphate C6-methyltransferase BezA binding with S-adenosylhomocysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BezA, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Tsutsumi, H, Moriwaki, Y, Terada, T, Shimizu, K, Katsuyama, Y, Ohnishi, Y. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and Molecular Basis of the Catalytic Mechanism of Geranyl Pyrophosphate C6-Methyltransferase: Creation of an Unprecedented Farnesyl Pyrophosphate C6-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1WTB
 
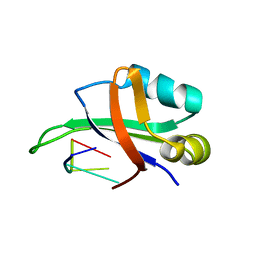 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D (AUF1) with telomere DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2004-11-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D
J.Biol.Chem., 280, 2005
|
|
